Abstract
All haematopoietic cell lineages that circulate in the blood of adult mammals derive from multipotent haematopoietic stem cells (HSCs)1. By contrast, in the blood of mammalian embryos, lineage-restricted progenitors arise first, independently of HSCs, which only emerge later in gestation2,3. As best defined in the mouse, ‘primitive’ progenitors first appear in the yolk sac at 7.5 days post-coitum2,3. Subsequently, erythroid–myeloid progenitors that express fetal haemoglobin4, as well as fetal lymphoid progenitors5, develop in the yolk sac and the embryo proper, but these cells lack HSC potential. Ultimately, ‘definitive’ HSCs with long-term, multilineage potential and the ability to engraft irradiated adults emerge at 10.5 days post-coitum from arterial endothelium in the aorta-gonad-mesonephros and other haemogenic vasculature3. The molecular mechanisms of this reverse progression of haematopoietic ontogeny remain unexplained. We hypothesized that the definitive haematopoietic program might be actively repressed in early embryogenesis through epigenetic silencing6, and that alleviating this repression would elicit multipotency in otherwise lineage-restricted haematopoietic progenitors. Here we show that reduced expression of the Polycomb group protein EZH1 enhances multi-lymphoid output from human pluripotent stem cells. In addition, Ezh1 deficiency in mouse embryos results in precocious emergence of functional definitive HSCs in vivo. Thus, we identify EZH1 as a repressor of haematopoietic multipotency in the early mammalian embryo.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Orkin, S. H. & Zon, L. I. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132, 631–644 (2008)
Medvinsky, A., Rybtsov, S. & Taoudi, S. Embryonic origin of the adult hematopoietic system: advances and questions. Development 138, 1017–1031 (2011)
Dzierzak, E. & Speck, N. A. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat. Immunol. 9, 129–136 (2008)
McGrath, K. E. et al. Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep. 11, 1892–1904 (2015)
Böiers, C. et al. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell 13, 535–548 (2013)
Cedar, H. & Bergman, Y. Epigenetics of haematopoietic cell development. Nat. Rev. Immunol. 11, 478–488 (2011)
Doulatov, S. et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell 13, 459–470 (2013)
Shen, X. et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502 (2008)
Laurenti, E. et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat. Immunol. 14, 756–763 (2013)
North, T. E. et al. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity 16, 661–672 (2002)
Rybtsov, S. et al. Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J. Exp. Med. 208, 1305–1315 (2011)
Boisset, J. C. et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120 (2010)
Riddell, J. et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 157, 549–564 (2014)
Papaemmanuil, E. et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat. Genet. 41, 1006–1010 (2009)
Tothova, Z. et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128, 325–339 (2007)
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006)
Xu, J. et al. Developmental control of Polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol. Cell 57, 304–316 (2015)
Margueron, R. et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503–518 (2008)
Jang, I. H. et al. Notch1 acts via Foxc2 to promote definitive hematopoiesis via effects on hemogenic endothelium. Blood 125, 1418–1426 (2015)
Hohmann, A. F. et al. Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat. Chem. Biol. 12, 672–679 (2016)
Müller, A. M., Medvinsky, A., Strouboulis, J., Grosveld, F. & Dzierzak, E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity 1, 291–301 (1994)
Yoder, M. C. et al. Characterization of definitive lymphohematopoietic stem cells in the day 9 murine yolk sac. Immunity 7, 335–344 (1997)
Lee, S. C. et al. Polycomb repressive complex 2 component Suz12 is required for hematopoietic stem cell function and lymphopoiesis. Blood 126, 167–175 (2015)
Xie, H. et al. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell 14, 68–80 (2014)
Majewski, I. J. et al. Polycomb repressive complex 2 (PRC2) restricts hematopoietic stem cell activity. PLoS Biol. 6, e93 (2008)
Kamminga, L. M. et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood 107, 2170–2179 (2006)
Cai, Z. et al. Haploinsufficiency of AML1 affects the temporal and spatial generation of hematopoietic stem cells in the mouse embryo. Immunity 13, 423–431 (2000)
Sugimura, R. et al. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature 545, 432–438 (2017)
Shao, Z., Zhang, Y., Yuan, G. C., Orkin, S. H. & Waxman, D. J. MAnorm: a robust model for quantitative comparison of ChIP–seq data sets. Genome Biol. 13, R16 (2012)
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010)
Cahan, P. et al. CellNet: network biology applied to stem cell engineering. Cell 158, 903–915 (2014)
Shin, H., Liu, T., Manrai, A. K. & Liu, X. S. CEAS: cis-regulatory element annotation system. Bioinformatics 25, 2605–2606 (2009)
Vo, L. T., Doulatov, S. & Daley, G. Q. Haematopoietic progenitor and lymphoid differentiation from human pluripotent stem cells. Protoc. Exch. http://doi.org/10.1038/protex.2017.160 (2018)
Park, I. H. et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146 (2008)
Chadwick, K. et al. Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood 102, 906–915 (2003)
Onder, T. T. et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature 483, 598–602 (2012)
Holmes, R. & Zúñiga-Pflücker, J. C. The OP9-DL1 system: generation of T-lymphocytes from embryonic or hematopoietic stem cells in vitro. Cold Spring Harb. Protoc. 2009, http://doi.org/10.1101/pdb.prot5156 (2009)
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013)
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008)
Huang, J. et al. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell 36, 9–23 (2016)
Kim, J., Cantor, A. B., Orkin, S. H. & Wang, J. Use of in vivo biotinylation to study protein–protein and protein–DNA interactions in mouse embryonic stem cells. Nat. Protoc. 4, 506–517 (2009)
Ditadi, A. et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat. Cell Biol. 17, 580–591 (2015)
Doulatov, S. et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 11, 585–593 (2010)
Acknowledgements
We thank T. Jenuwein for sharing the Ezh1 mutant mice, which were generated at the Research Institute of Molecular Pathology (IMP, Vienna) in 2000 by D. O’Carroll (laboratory of T. Jenuwein) with the help of M. Sibilia (laboratory of E. Wagner). We also thank T. Schlaeger and the hESC Core Facility at Boston Children’s Hospital for providing pluripotent stem-cell lines, R. Mathieu from BCH Flow Cytometry Core, and M. J. Chen for technical advice. This work was supported by grants from the NIH NIDDK (R24-DK092760, R24-DK49216) and NHLBI Progenitor Cell Biology Consortium (U01-HL100001); NHLBI R01HL04880 and NIH R24OD017870-01. L.T.V. is supported by the NSF Graduate Research Fellowship. M.A.K. is supported by T32 NIH Training Grant from BWH Hematology. M.C. is supported by a fellowship from the Leukemia and Lymphoma Society. S.D. is supported by K99 NIH NHLBI award (1K99HL123484). S.H.O. is an Investigator of the Howard Hughes Medical Institute. J.X. is supported by NIH grants (K01DK093543 and R01DK111430) and a Cancer Prevention and Research Institute of Texas (CPRIT) New Investigator award (RR140025). G.Q.D. was supported by the Howard Hughes Medical Institute, and is an associate member of the Broad Institute and an investigator of the Manton Center for Orphan Disease Research.
Author information
Authors and Affiliations
Contributions
L.T.V., S.D. and G.Q.D. conceived the project. L.T.V. designed all experiments, performed all pluripotent stem-cell and mouse transplantation studies and interpreted data. M.A.K. analysed RNA-seq, ChIP–seq and ATAC–seq data, performed all network analyses and interpreted data. X.L. performed ChIP–seq and ATAC–seq experiments. Y.Z. and Z.S. analysed ChIP–seq and ATAC–seq data. J.B. performed and analysed qPCR and western blot validations, assisted with tissue culture, animal dissections and mouse transplantation studies. P.M.S. assisted with timed matings, animal dissections and mouse transplantation studies. D.K.J. performed western blot validations, cloned the Ezh2-mCherry overexpression construct, assisted with ChIP–seq optimization and interpreted data. M.C. assisted with ChIP–seq optimization. A.H. assisted with RNA-seq analysis. T.E.N., S.H.O., S.D., J.X. and G.Q.D. supervised research, interpreted data and participated in project planning. L.T.V., T.E.N., S.D. and G.Q.D. wrote the manuscript with input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks B. Gottgens, H. Mikkola and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
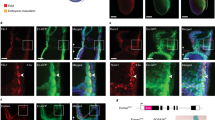
Extended Data Figure 1 EZH1 knockdown activates lymphoid potential from pluripotent stem cells.
a, List of all candidate epigenetic modifiers in loss-of-function shRNA screen. b, Representative flow plots of CD4+CD8+ T cell potential across top six candidates from four independent hairpins in two independent experiments (n = 8). See Fig. 1. c, CD34+ cells were isolated after 9 days of embryoid body (EB) differentiation (top left), transduced with shLUC or shEZH1 and cultured under conditions that promote endothelial-to-haematopoietic transition44. After 6 days, rounded haematopoietic cells (top right) were collected and co-cultured on OP9-DL1 stroma. Bottom, flow cytometric analysis of T cell potential in shLUC and shEZH1 cells without 5F is shown for two independent iPS lines (34-iPS and MSC-iPS1) in one experiment (n = 2 biological replicates). PSC-HE, pluripotent stem-cell-derived haemogenic endothelium. d, Expansion and differentiation potential of 5F plus shEZH1 cells after long-term in vitro culture. 5F plus shEZH1 cells were maintained in cultures containing doxycycline for 14 days respecification (approximately 100-fold expansion), plus an additional 6 weeks (approximately 1,000-fold expansion) and then plated into OP9-DL1 stromal cells for T cell differentiation. Representative flow cytometric analyses of T cell potential of 5F plus shLUC and 5F plus shEZH1 cells after 13 weeks of expansion and differentiation (n = 2 biological replicates). e, Flow cytometric analysis (left) and quantification (right) of the proportion of CD34+ and CD34− haematopoietic progenitors in doxycycline-containing suspension culture at day 25 (n = 2 biological replicates).
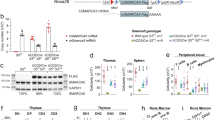
Extended Data Figure 2 Ezh1, but not Ezh2, suppresses T cell potential and requires its catalytic domain.
a, qPCR of PRC2 expression (human genes EZH1, EED, EZH2 and SUZ12), during the course of differentiation from human pluripotent stem cell-derived CD34+ cells, respecification (5F), expansion, OP9-DL1 co-culture and CD4+CD8+ T cells (n = 2 biological replicates in one experiment). b, qPCR of mRNA knockdown efficiency of individual shRNAs for PRC2 genes (n = 2 biological replicates). See also Fig. 2a, b. c, Western blot for EZH1 and GAPDH protein levels. d, Scheme for rescue experiments. GFP+ 5F cells were transduced with shRNAs and selected with puromycin. 5F plus shRNA cells were then transduced with full-length mouse Ezh1 open-reading frame (mEzh1) or mutant mouse Ezh1 with the catalytic SET domain deleted (mEzh1ΔSET), marked by mCherry fluorescence. Triple-transduced (GFP+, puromycin-resistant, mCherry+) cells were sorted and seeded onto OP9-DL1. T cells were analysed by flow cytometry after 5 weeks of differentiation. See also Fig. 2c. e, Expression of full-length mouse Ezh1, catalytic-deleted mEzh1ΔSET, or full-length mouse Ezh2 in shLUC and shEZH1 cells by qPCR. f, Western blot validation of expression of mouse Ezh1 or mutant mEzh1ΔSET in shLUC and shEZH1 cells. g, Top, representative flow cytometry plots of T cell potential for 5F plus shLUC cells for rescue experiments in d (n = 3 biological replicates). Bottom, CD4+CD8+ T cells were verified for mCherry fluorescence. See also Fig. 2c. All plots are gated on CD45+. h, 5F plus shRNA cells were transduced with full-length mouse Ezh2 open-reading frame (mEzh2) marked by mCherry fluorescence. Triple-transduced (GFP+, puromycin-resistant, mCherry+) cells were sorted and seeded onto OP9-DL1 stromal cells. T cells were analysed by flow cytometry after 5 weeks of differentiation. Representative flow plots for two biological replicates in one experiment. i, Quantification of data in h. Data are mean ± s.e.m.
Extended Data Figure 3 Ezh1 regulates haematopoietic and lymphoid programs in vitro and in vivo.
a, Representative images of E10.5 embryo (top), yolk sac (middle) and AGM (bottom) from n > 30 embryos. Lin−c-Kit+VE-cadherin+CD45+CD41+ cells from E10.5 yolk sac and AGM were FACS-sorted followed by RNA-seq analysis. See also Fig. 4a, c, d. b, Genes upregulated and downregulated by more than twofold in Ezh1+/− or Ezh1−/− yolk sac and AGM compared to those from wild-type mice. c, d, GO term annotations of upregulated genes in Ezh1+/− and Ezh1−/− yolk sac and AGM compared to those from wild-type mice. e, GO analysis of enriched pathways of the 1,033 nearest neighbour genes associated with upregulated ATAC peaks (top) and the nearest 1,012 neighbour genes associated with downregulated ATAC peaks (bottom). See also Fig. 3d, e. f, Comparison of upregulated ATAC peaks in 5F plus shEZH1 cells with HSPC hierarchy signatures45 (top) and HSPC B and T cell networks (bottom). See also Fig. 3d, e. g, Box plot of expression of genes associated with upregulated and downregulated ATAC peaks. *P < 0.05 by one-way ANOVA. h, ATAC density map of c-Kit+VE-cadherin+CD45+ HSPCs sorted from approximately 30 embryos of E10.5 wild-type and Ezh1−/− AGM (top) from one experiment. Significantly upregulated ATAC peaks were compared to HSPC, T, B cell networks and signatures of the human HSPC hierarchy (bottom). See also Fig. 4c, d. i, GO terms of enriched pathways of regions associated with significantly upregulated ATAC peaks annotated by GREAT analysis in Ezh1+/− AGM (top) and Ezh1−/− AGM (bottom) compared to wild type. See also Fig. 4c, d. j, GO terms of enriched pathways of regions associated with significantly downregulated ATAC peaks annotated by GREAT analysis30 in Ezh1+/− AGM (top) and Ezh1−/− AGM (bottom) compared to wild type. See also Fig. 4c. k, Transcription factor binding to genes with upregulated ATAC peaks in Ezh1+/− (left) and Ezh1−/− (right) AGM from i compared to wild-type AGM.
Extended Data Figure 4 Genome-wide chromatin occupancy reveals EZH1 enrichment at bivalent HSC genes and non-canonical active lymphoid genes.
a, Breakdown of EZH1 binding at promoter regions and associated histone marks. b–d, GO term analysis of EZH1-bound active (b), bivalent (c) and repressed (d) genes. e, Distribution of EZH1-bound genes across the haematopoietic hierarchy (left) and their associated histone marks (right). A, active (H3K4me3-marked); B, bivalent (H3K4me3 and H3K27me3-marked); R, repressed (H3K27me3-marked). f, GSEA analysis of EZH1-bound genes correlated with RNA-seq data upon EZH1 knockdown. g, Sankey diagram showing genome-wide changes in histone methylation status upon EZH1 knockdown. h, Upregulated genes exhibit reciprocal decreases in H3K27me3 levels, as quantified by EpiChIP software. K4, H3K4me3; K27, H3K27me3. See also Fig. 3i. i, Activated (formerly bivalent) HSC genes exhibit increased gene expression upon EZH1 knockdown and loss of H3K27me3. See also Fig. 3j. j, Correlations between changes in H3K27me3 and gene expression levels upon EZH1 knockdown, subdivided by subgroups corresponding to methylation changes. N, null. k, Breakdown of bivalent–bivalent (left), bivalent–repressed (centre) and bivalent–null (right) genes upon EZH1 knockdown across the haematopoietic hierarchy. l, Overlap of EZH1- and EZH2-enriched peaks and the distribution of all EZH1-enriched, EZH2-enriched or common genes across the hierarchy (left), or specifically bivalent genes that become activated upon EZH1 knockdown (middle) and active genes, marked by H3K4me3 in shLUC (right). m, SUZ12 binding (from the ChEA database) across the haematopoietic hierarchy. n, Canonical and non-canonical previously identified targets17 across the haematopoietic hierarchy. o, p, Breakdown of histone marks on non-canonical ProB genes (o) and the genome-wide distribution from CEAS analysis32 (p). q, Changes in actively marked, non-canonical ProB genes (green bar in o), upon EZH1 knockdown. r, SUZ12 and EZH2 binding (ChEA database) at ATAC peaks in Ezh1+/− and Ezh1−/− AGM. *P < 0.05 by one-way ANOVA.
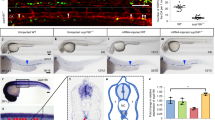
Extended Data Figure 5 Ezh1 deficiency enhances embryonic HSPC engraftment.
a, Whole E10.5 AGM and yolk sac were transplanted intravenously into sublethally irradiated NSG adult females. Chimaerism was monitored by retroorbital bleeding every 4 weeks. Representative flow plots are shown for analysis after 4 weeks in n ≥ 3 mice. See also Fig. 4e, f. b, Whole E9.5 PSP was transplanted intravenously into sublethally irradiated NSG adult females. Chimaerism was monitored via retroorbital bleeding every 4 weeks. Representative flow plots are shown for analysis after 8 weeks in n ≥ 3 mice. See also Fig. 4g. c, Representative flow plots of lineage analysis in E10.5 AGM Ezh1+/− and Ezh1−/− primary transplant recipients after 24 weeks, and in E9.5 PSP Ezh1+/− primary transplant recipient after 16 weeks (n ≥ 3 mice per group). See also Fig. 4e, g. d, Primary recipients in a were euthanized 24 weeks post-transplantation and 4 × 106 whole bone marrow was transplanted into sublethally irradiated adult NSG females intravenously. Chimaerism was monitored via retroorbital bleeding. Representative flow plots of E10.5 AGM and yolk sac secondary transplants after 4 weeks in n ≥ 3 mice. See also Fig. 4h. e, Left, secondary transplantation of E10.5 yolk sac primary recipients (Fig. 4f). Right, lineage distribution of E10.5 secondary recipients. Data are pooled across three independent experiments. *P < 0.05, ** P < 0.01 by unpaired two-sided t-test; see Supplementary Information for exact P values per time point.
Extended Data Figure 6 Ezh1-deficient embryonic HSPCs contribute to adult-type lymphopoiesis in vivo.
a, Flow analysis of B1 and B2 progenitors in the peritoneal cavity of engrafted primary recipients (n = 1 mouse per group). b, Flow analysis of TCRβ and TCRγδ frequencies of donor-derived peripheral CD3+ T cells from engrafted primary recipients (n = 1 mouse per group). See also Fig. 4e, f.
Supplementary information
Supplementary Information
This file contains the uncropped gels and a list of sequences used in the manuscript. (PDF 1176 kb)
Rights and permissions
About this article
Cite this article
Vo, L., Kinney, M., Liu, X. et al. Regulation of embryonic haematopoietic multipotency by EZH1. Nature 553, 506–510 (2018). https://doi.org/10.1038/nature25435
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25435
This article is cited by
-
Gain and loss of function variants in EZH1 disrupt neurogenesis and cause dominant and recessive neurodevelopmental disorders
Nature Communications (2023)
-
Generation and clinical potential of functional T lymphocytes from gene-edited pluripotent stem cells
Experimental Hematology & Oncology (2022)
-
Evolutionary adaptation of the Polycomb repressive complex 2
Epigenetics & Chromatin (2022)
-
Integrative epigenomic and transcriptomic analysis reveals the requirement of JUNB for hematopoietic fate induction
Nature Communications (2022)
-
Continuous human iPSC-macrophage mass production by suspension culture in stirred tank bioreactors
Nature Protocols (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.