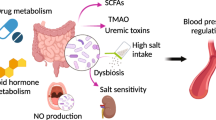
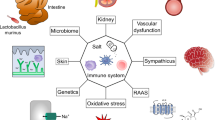
Abstract
A Western lifestyle with high salt consumption can lead to hypertension and cardiovascular disease. High salt may additionally drive autoimmunity by inducing T helper 17 (TH17) cells, which can also contribute to hypertension. Induction of TH17 cells depends on gut microbiota; however, the effect of salt on the gut microbiome is unknown. Here we show that high salt intake affects the gut microbiome in mice, particularly by depleting Lactobacillus murinus. Consequently, treatment of mice with L. murinus prevented salt-induced aggravation of actively induced experimental autoimmune encephalomyelitis and salt-sensitive hypertension by modulating TH17 cells. In line with these findings, a moderate high-salt challenge in a pilot study in humans reduced intestinal survival of Lactobacillus spp., increased TH17 cells and increased blood pressure. Our results connect high salt intake to the gut–immune axis and highlight the gut microbiome as a potential therapeutic target to counteract salt-sensitive conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Manzel, A. et al. Role of “Western diet” in inflammatory autoimmune diseases. Curr. Allergy Asthma Rep. 14, 404 (2014)
O’Donnell, M. et al. Urinary sodium and potassium excretion, mortality, and cardiovascular events. N. Engl. J. Med. 371, 612–623 (2014)
Weber, M. A. et al. Clinical practice guidelines for the management of hypertension in the community: a statement by the American Society of Hypertension and the International Society of Hypertension. J. Hypertens. 32, 3–15 (2014)
Taylor, J. 2013. ESH/ESC guidelines for the management of arterial hypertension. Eur. Heart J. 34, 2108–2109 (2013)
Mozaffarian, D. et al. Global sodium consumption and death from cardiovascular causes. N. Engl. J. Med. 371, 624–634 (2014)
Coffman, T. M. Under pressure: the search for the essential mechanisms of hypertension. Nat. Med. 17, 1402–1409 (2011)
Wenzel, U. et al. Immune mechanisms in arterial hypertension. J. Am. Soc. Nephrol. 27, 677–686 (2016)
Guzik, T. J. et al. Role of the T cell in the genesis of angiotensin II-induced hypertension and vascular dysfunction. J. Exp. Med. 204, 2449–2460 (2007)
Madhur, M. S. et al. Interleukin 17 promotes angiotensin II-induced hypertension and vascular dysfunction. Hypertension 55, 500–507 (2010)
Norlander, A. E. et al. Interleukin-17A regulates renal sodium transporters and renal injury in angiotensin II-induced hypertension. Hypertension 68, 167–174 (2016)
Kleinewietfeld, M. et al. Sodium chloride drives autoimmune disease by the induction of pathogenic TH17 cells. Nature 496, 518–522 (2013)
Wu, C. et al. Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature 496, 513–517 (2013)
Miossec, P., Korn, T. & Kuchroo, V. K. Interleukin-17 and type 17 helper T cells. N. Engl. J. Med. 361, 888–898 (2009)
Bettelli, E. Building different mouse models for human MS. Ann. NY Acad. Sci. 1103, 11–18 (2007)
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014)
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006)
Honda, K. & Littman, D. R. The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84 (2016)
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009)
Cole, J. R. et al. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2013)
Lozupone, C. & Knight, R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71, 8228–8235 (2005)
Sun, Z. et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 6, 8322 (2015)
Wannemuehler, M. J., Overstreet, A. M., Ward, D. V. & Phillips, G. J. Draft genome sequences of the altered Schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announc. 2, e00287–e14 (2014)
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014)
Zelante, T. et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39, 372–385 (2013)
Jangi, S. et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 7, 12015 (2016)
Berer, K. et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479, 538–541 (2011)
Haghikia, A. et al. Dietary fatty acids directly impact central nervous system autoimmunity via the small intestine. Immunity 43, 817–829 (2015)
Rothhammer, V. et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 (2016)
Khalesi, S., Sun, J., Buys, N. & Jayasinghe, R. Effect of probiotics on blood pressure: a systematic review and meta-analysis of randomized, controlled trials. Hypertension 64, 897–903 (2014)
Hansen, T. W. et al. Predictive role of the nighttime blood pressure. Hypertension 57, 3–10 (2011)
Forslund, K. et al. Country-specific antibiotic use practices impact the human gut resistome. Genome Res. 23, 1163–1169 (2013)
Voigt, A. Y. et al. Temporal and technical variability of human gut metagenomes. Genome Biol. 16, 73 (2015)
Mende, D. R., Sunagawa, S., Zeller, G. & Bork, P. Accurate and universal delineation of prokaryotic species. Nat. Methods 10, 881–884 (2013)
Farez, M. F., Fiol, M. P., Gaitán, M. I., Quintana, F. J. & Correale, J. Sodium intake is associated with increased disease activity in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 86, 26–31 (2015)
Fitzgerald, K. C. et al. Sodium intake and multiple sclerosis activity and progression in BENEFIT. Ann. Neurol. 82, 20–29 (2017)
Lamas, B. et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605 (2016)
Wang, Y. et al. Kynurenine is an endothelium-derived relaxing factor produced during inflammation. Nat. Med. 16, 279–285 (2010)
Binger, K. J. et al. High salt reduces the activation of IL-4- and IL-13-stimulated macrophages. J. Clin. Invest. 125, 4223–4238 (2015)
Eil, R. et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature 537, 539–543 (2016)
Hernandez, A. L. et al. Sodium chloride inhibits the suppressive function of FOXP3+ regulatory T cells. J. Clin. Invest. 125, 4212–4222 (2015)
Jantsch, J. et al. Cutaneous Na+ storage strengthens the antimicrobial barrier function of the skin and boosts macrophage-driven host defense. Cell Metab. 21, 493–501 (2015)
Cox, L. M. et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158, 705–721 (2014)
Tamburini, S., Shen, N., Wu, H. C. & Clemente, J. C. The microbiome in early life: implications for health outcomes. Nat. Med. 22, 713–722 (2016)
Martínez, I. et al. The gut microbiota of rural Papua New Guineans: composition, diversity patterns, and ecological processes. Cell Reports 11, 527–538 (2015)
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012)
Preheim, S. P., Perrotta, A. R., Martin-Platero, A. M., Gupta, A. & Alm, E. J. Distribution-based clustering: using ecology to refine the operational taxonomic unit. Appl. Environ. Microbiol. 79, 6593–6603 (2013)
Edgar, R. C. & Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482 (2015)
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007)
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010)
Oksanen, J . et al. Vegan: Community Ecology Package. R package v.2.3–0 http://CRAN.R-project.org/package=vegan (2015)
Caporaso, J. G. et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26, 266–267 (2010)
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011)
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010)
Paradis, E., Claude, J. & Strimmer, K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004)
Freund, Y. & Schapire, R. E. A decision-theoretic generalization of on-line learning and an application to boosting. J. Comput. Syst. Sci. 55, 119–139 (1997)
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001)
Pietzke, M., Zasada, C., Mudrich, S. & Kempa, S. Decoding the dynamics of cellular metabolism and the action of 3-bromopyruvate and 2-deoxyglucose using pulsed stable isotope-resolved metabolomics. Cancer Metab. 2, 9 (2014)
Kuich, P. H., Hoffmann, N. & Kempa, S. Maui-VIA: a user-friendly software for visual identification, alignment, correction, and quantification of gas chromatography–mass spectrometry data. Front. Bioeng. Biotechnol. 2, 84 (2015)
Hartemink, R., Domenech, V. R. & Rombouts, F. M. LAMVAB—A new selective medium for the isolation of lactobacilli from faeces. J. Microbiol. Methods 29, 77–84 (1997)
Gomila, M. et al. Genotypic and phenotypic applications for the differentiation and species-level identification of Achromobacter for clinical diagnoses. PLoS One 9, e114356 (2014)
Itani, H. A. et al. CD70 exacerbates blood pressure elevation and renal damage in response to repeated hypertensive stimuli. Circ. Res. 118, 1233–1243 (2016)
Atarashi, K. & Honda, K. Analysis of murine lamina propria TH17 cells. Protoc. Exch. http://doi.org/10.1038/nprot.2008.205 (2008)
Wiig, H. et al. Immune cells control skin lymphatic electrolyte homeostasis and blood pressure. J. Clin. Invest. 123, 2803–2815 (2013)
Mähler, A. et al. Increased catabolic state in spinocerebellar ataxia type 1 patients. Cerebellum 13, 440–446 (2014)
Sunagawa, S. et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat. Methods 10, 1196–1199 (2013)
Kultima, J. R. et al. MOCAT2: a metagenomic assembly, annotation and profiling framework. Bioinformatics 32, 2520–2523 (2016)
Segata, N. et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814 (2012)
Therneau, T. M. A package for survival analysis in S. v.2.38 https://CRAN.R-project.org/package=survival (2015)
Therneau, T. M. & Grambsch, P. M. Modeling Survival Data: Extending the Cox Model (Springer, 2000)
Yuan, S., Cohen, D. B., Ravel, J., Abdo, Z. & Forney, L. J. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One 7, e33865 (2012)
Caesar, R., Tremaroli, V., Kovatcheva-Datchary, P., Cani, P. D. & Bäckhed, F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 22, 658–668 (2015)
Bergström, A. et al. Introducing GUt low-density array (GULDA): a validated approach for qPCR-based intestinal microbial community analysis. FEMS Microbiol. Lett. 337, 38–47 (2012)
Bindels, L. B. et al. Restoring specific lactobacilli levels decreases inflammation and muscle atrophy markers in an acute leukemia mouse model. PLoS One 7, e37971 (2012)
Duniere, L. et al. Impact of adding Saccharomyces strains on fermentation, aerobic stability, nutritive value, and select lactobacilli populations in corn silage. J. Anim. Sci. 93, 2322–2335 (2015)
Byun, R. et al. Quantitative analysis of diverse Lactobacillus species present in advanced dental caries. J. Clin. Microbiol. 42, 3128–3136 (2004)
Cui, Y. et al. Different effects of three selected Lactobacillus strains in dextran sulfate sodium-induced colitis in BALB/c mice. PLoS One 11, e0148241 (2016)
Matsuda, K. et al. Establishment of an analytical system for the human fecal microbiota, based on reverse transcription–quantitative PCR targeting of multicopy rRNA molecules. Appl. Environ. Microbiol. 75, 1961–1969 (2009)
Acknowledgements
We thank G. N’diaye, I. Kamer, S. Seubert, P. Voss, J. Anders, C. Schmidt, A. Geuzens, R. Hercog and S. Kandels-Lewis for assistance; and J. J. Mullins and F. C. Luft for their support. This study was funded by grants from the German Centre for Cardiovascular Research (DZHK; BER 1.1 VD), the Center for Microbiome Informatics and Therapeutics, and the MetaCardis consortium. D.N.M., J.J. and M.G. were supported by the German Research Foundation (DFG). R.A.L. holds an endowed professorship supported by Novartis Pharma. M.K. was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (640116), by a SALK-grant from the government of Flanders, Belgium and by an Odysseus-grant of the Research Foundation Flanders (FWO), Belgium. L. reuteri was provided by L. Romani.
Author information
Authors and Affiliations
Contributions
N.W. led and conceived the project, designed and performed most experiments, analysed and interpreted the data. M.G.M., S.W.O. and S.M.K. performed 16S sequencing and data analysis. S.H., D.T., M.Ba. and C.W. performed animal experiments and analysed data. H.B., S.H., A.B., D.A.G. and B.F.C.-R. performed and analysed flow cytometry. L.Mai., S.M.K., V.S., P.N. and R.G.G. performed bacterial growth experiments. O.V. and C.F. performed metabolite analysis with input from A.B. and M.G.M. L.Mar., F.H.K. and L.K. performed 16S qPCR. N.R. performed sodium analyses. K.F. performed metagenomic analyses with contributions from P.I.C. and S.S. M.Bo., R.D. and A.M. conducted the clinical study. A.K. performed statistical analyses. M.G., A.T., J.M.T., S.K., P.B. and J.J. supervised the experiments and analyses. D.N.M., E.J.A., M.K. and R.A.L. conceived the project, supervised the experiments and interpreted the data. N.W. and D.N.M. wrote the manuscript with key editing by E.J.A., R.A.L., M.K. and K.F. and further input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks T. Coffman, D. A. Relman, H. Wekerle and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Body weight, food, fluid and sodium chloride (NaCl) intake, and intestinal transit in mice fed a NSD or HSD.
a–f, Body weight (a), food intake (b), fluid intake (c), NaCl intake from the chow (d), NaCl intake from the drinking water (e) and total NaCl intake (f; sum of NaCl intake from chow and drinking water) in mice fed a NSD (n = 8) or HSD (n = 8). g, Measurement of intestinal transit. FVB/N mice were fed a NSD (n = 8) or HSD (n = 9) for 14 days and administered activated charcoal (0.5 g per 10 ml in 0.5% methylcellulose; 0.1 ml per 10 g body weight by oral gavage). After 20 min, mice were euthanized and the distance travelled by charcoal was measured. Data are mean ± s.e.m., circles represent individual mice. **P < 0.01; ****P < 0.001; paired two-tailed Student’s t-test (a–c), one-tailed Wilcoxon matched-pairs signed-rank test (d–f), unpaired two-tailed Student’s t-test (g).
Extended Data Figure 2 Faecal microbiome profiles of mice kept on a NSD or HSD over time.
Taxonomic bar charts showing relative abundance of ribosomal database project-based OTUs on indicated days. a, Mice remaining on a NSD for 14 days served as NSD controls. Baseline NSD day −1 and NSD day 14 are shown. b, Separate mice were switched from NSD (days −2 and −1) to HSD for 14 days, and were finally re-exposed to NSD for another 14 days (recovery). For time course analyses, faecal samples from baseline NSD days (−1 and −2), early (days 1–3) and late (day 14) HSD days and NSD recovery days (days 15–17, 19, 22, 28) are shown. n = 8 mice per group.
Extended Data Figure 3 HSD alters the faecal microbiome and the faecal metabolite profile.
a, Mouse 16S rDNA faecal microbiome samples do not separate by diet in a MDS ordination (white, NSD samples; black, HSD samples; grey, recovery on NSD). b, qPCR of DNA extracted from faecal samples of mice fed a NSD or HSD using universal 16S rDNA primers (n = 8 faecal samples per group from independent mice, indicated by circles; two-tailed Wilcoxon matched-pairs signed-rank test). c, Phylogenetic tree showing changes in microbiome composition caused by HSD. OTUs present in samples from day 14 are indicated by coloured circles (red indicates reduction in HSD samples; blue indicates enrichment). The radius of each circle indicates a median log-fold difference in relative abundance between the two diets. Filled circles mark statistically significant differences (two-tailed Student’s t-test, Benjamini–Hochberg correction, P < 0.05). d–g, High dietary salt strongly influences the faecal metabolite profile. Male FVB/N mice (n = 8) were fed a NSD and then switched to HSD. Metabolites were extracted from faecal pellets taken from mice on a NSD (day −3) and HSD (day 13), and analysed by GC–MS. d, HSD samples are clearly distinguishable from NSD samples in a principal component analysis for faecal metabolites. e, Faecal metabolites clearly cluster by treatment. The majority of faecal metabolites are reduced by HSD. Hierarchically clustered heat map, metabolites shown in alphabetical order. Metabolites were normalized by subtracting the minimum and dividing by the maximum value across all mice. f, Faecal levels of the nucleoside adenosine were similar in both diets, suggesting that the change in metabolites is not due to a decrease in overall bacterial biomass. g, HSD leads to a reduction in total metabolite peak intensities in faecal samples. **P < 0.01; two-tailed paired Student’s t-test (f, g).
Extended Data Figure 4 Accuracy of AdaBoost and random forest classifiers.
a, AdaBoost and random forest classifiers (trained on samples from days −2, −1 and 14) were used to predict the classification of all samples from HSD mice. The fraction of samples from each time point that the classifiers predicted as belonging to animals currently on a HSD is shown. The two runs of the random forest produced the same fractions, so only one line is shown for the two random forest classifiers. b, Time series for the remaining 7 OTUs important to the classifier. NSD and HSD phases are indicated by white and grey backgrounds. Mice (n = 12) were switched from NSD to HSD and back to NSD (subgroup of n = 8). Other control mice (n = 8) that remaind on NSD are shown in white. Box plots show median, IQR with whiskers of 1.5× IQR, circles represent samples from independent mice.
Extended Data Figure 5 L. murinus genome and in vitro growth of Lactobacilli.
a, Venn diagram of the coding sequences present in the L. murinus and two other isolates with available with full genome sequences. b, Bootstrapped phylogenetic tree of full-length 16S rDNA from a variety of Lactobacillus species resident in rodent or human guts. Prevalence of the respective species in the MetaHIT cohort is shown. L. murinus strains are absent from the MetaHIT cohort. c, Growth yield (OD600) of L. murinus measured at increasing concentrations of NaCl. Aerobic endpoint measurements of liquid L. murinus cultures in MRS medium and increasing NaCl concentrations relative to growth in MRS without the addition of NaCl. n = 5 independent experiments. d, Anaerobic growth yield of L. murinus, A. muciniphila, P. excrementihominis and C. difficile grown at 37 °C for 48 h in MGAM liquid medium. Growth at each salt concentration is normalized to growth at 0.086 M Na+. The respective IC50 is indicated. n = 3 technical replicates across two experiments. e, Anaerobic growth of selected human Lactobacillus species in MGAM medium with increasing NaCl concentrations. Relative growth yield is calculated based on AUCs by comparing to growth in MGAM without the addition of NaCl. n = 3 independent experiments with three technical replicates. f, Heat map showing data as in e. The respective IC50 is shown in the bottom row. g, Growth yield of E. coli and L. murinus, grown at 37 °C for 12–16 h in LB (E. coli) or MRS broth (L. murinus). n = 4 technical replicates from two independent experiments. Data are mean ± s.e.m.
Extended Data Figure 6 Indole metabolites in mouse faecal samples.
a, b, Effect of HSD on faecal IAA (a) and IAld (b) content in FVB/N mice fed a NSD or HSD (n = 12 per group in a; n = 13 per group in b). c, d, Germ-free (GF) mice monocolonized with L. murinus showed increased faecal IAA and IAld content (n = 8 per group). *P < 0.05; ****P < 0.0001; one-tailed Wilcoxon matched-pairs signed-rank test (a, b), one-tailed Mann–Whitney U-test (c), unpaired one-tailed Student’s t-test (d). n represents the number of independent mice.
Extended Data Figure 7 The effect of Lactobacillus species on actively induced EAE.
a, Median cumulative clinical EAE scores at day 15, 16 and 17 post immunization (p.i.) of NSD (n = 9), HSD (n = 11) and HSD mice treated with L. murinus (n = 6) starting at the day of immunization. b, Clinical course of MOG35–55 EAE in NSD mice (black circles, n = 7) and NSD mice treated with L. murinus (green squares, n = 4). c, Clinical course of MOG35–55 EAE in HSD mice (black circles) and HSD mice treated with L. reuteri (green squares, n = 6 independent mice per group). d, e, Quantification of CD4+IL-17A+IFNγ− cells on day 17 of EAE in the spleen (d) and spinal cord (e). n = 4 independent mice per group. f–h, Spinal cords on day 17 of EAE were analysed by RT–qPCR for relative expression of Il17a (f; n = 7 for NSD, n = 6 for HSD and n = 5 for HSD with L. murinus), Rorc (g; n = 5 for NSD, n = 6 for HSD and n = 5 for HSD with L. murinus) and Csf2 (h; n = 8 for NSD, n = 6 for HSD and n = 4 for HSD with L. murinus). i, Quantification of IFNγ-producing TH1 cells of siLPL on day 3 of EAE (n = 4 per group) and quantification of IFNγ-producing TH1 cells in spleen (n = 4 per group) and spinal cord on day 17 of EAE (n = 5 for NSD, n = 6 for HSD and n = 5 for HSD with L. murinus). j–m, Faecal indole metabolites were determined in MOG35–55 EAE mice by LC–MS/MS analysis. Effect of HSD on faecal IAA (j) and IAld (k) content on day 10 after immunization (n = 5 per group (j), n = 4 for NSD and n = 5 for HSD (k)). Faecal IAA (l) and IAld (m) content in MOG35–55 EAE mice fed a HSD with or without concomitant L. murinus treatment on day 10 after immunization (n = 7 per group (l), n = 8 for HSD and n = 7 for HSD with L. murinus for (m)). Box plots show median and IQR, whiskers are minimum and maximum values (a, j–m), data are mean ± s.e.m. b–i). *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant; Kruskal–Wallis test followed by Dunn’s multiple comparisons test (a), two-tailed Mann–Whitney U-test (b, c), one-tailed Mann–Whitney U test (d, e), one-way ANOVA followed by Tukey’s post hoc test (f–h), one-way ANOVA (i), unpaired one-tailed Student’s t-test (j, l, m), one-tailed Wilcoxon matched-pairs signed-rank test (k). n represents the number of independent mice per group, indicated by circles.
Extended Data Figure 8 Actively induced EAE in gnotobiotic mice.
a, b, HSD fails to induce intestinal TH17 cells in germ-free MOG35–55 EAE mice (n = 5 for GF + NSD and n = 6 for GF + HSD). a, Analysis of IL-17A and IFNγ in CD4+ siLPL isolated from NSD or HSD-fed MOG35–55-immunized germ-free mice (day 3 after immunization). Representative flow cytometry plots (left) show one mouse per group. Quantifications show frequencies of CD4+IL-17A+IFNγ− (middle) and CD4+IL-17A+IFNγ+ (right) cells. b, Quantification of CD4+RORγt+ frequencies in siLPL. c, d, L. murinus reduces small intestinal (siLPL) and colonic (cLPL) lamina propria TH17 cells in EAE mice colonized with SFB. MOG35–55 EAE was induced in germ-free mice monocolonized with SFB (GF + SFB) and germ-free mice colonized with SFB and L. murinus (GF + SFB + L. murinus). LPL were isolated on day 3 after immunization. c, Left, representative flow cytometry plots demonstrating IL-17A and IFNγ expression in CD4+ siLPL (one mouse per group). Middle, quantification of CD4+IL-17A+IFNγ− siLPL (n = 9 for GF + SFB, n = 8 for GF + SFB + L. murinus). Right, quantification of CD4+RORγt+ siLPL (n = 9 mice per group). d, Left, representative flow cytometry plots (one mouse per group) depicting IL-17A and IFNγ expression in CD4+ cLPL. Middle and right, quantification of CD4+IL-17A+IFNγ− (n = 8 for GF + SFB, n = 9 for GF + SFB + L. murinus) and CD4+IL-17A+IFNγ+ cLPL (n = 8 per group). Data are mean ± s.e.m., circles represent independent mice. *P < 0.05; ***P < 0.001; unpaired one-tailed Student’s t-test (a–d).
Extended Data Figure 9 Treatment with L. murinus or L. reuteri ameliorates salt-sensitive hypertension.
a, Mean diastolic pressures over time in response to HSD and HSD with concomitant L. murinus treatment in n = 7 FVB/N mice. Scale bar indicates 24 h. Horizontal line indicates the mean across all values of the respective phase. b, c, Mean systolic (b) and diastolic (c) blood pressures in mice (n = 7) fed a HSD (black curve) and HSD with concomitant L. murinus treatment at circadian scale. Arrows indicate the time of L. murinus gavage. d, e, Box plots (median, IQR, whiskers 1.5× IQR) show systolic (d) and diastolic (e) blood pressures recorded continuously in FVB/N mice fed a HSD and a HSD with concomitant L. reuteri treatment. These mice (n = 9) were fed a HSD for 10 days before concomitant L. reuteri treatment for another 7 days. ***P < 0.001 versus HSD using a linear mixed model. f, g, Box plots (median, IQR, whiskers 1.5× IQR) show systolic (f) and diastolic (g) blood pressures in mice (n = 5) fed a HSD and a HSD with concomitant E. coli Nissle 1917 treatment for three days, respectively. Statistics using linear mixed model. h–j, Quantification of CD4+IL-17A−IFNγ+ lymphocytes in siLPL (h; n = 5 for NSD, n = 7 for HSD, n = 6 for HSD + L. murinus) and cLPL and spleen, respectively (i, j, n = 5 for NSD, n = 6 for HSD, n = 6 for HSD + L. murinus). Data are mean ± s.e.m., circles represent independent mice. *P < 0.05; Kruskal–Wallis and Dunn’s post hoc test (h), one-way ANOVA (i, j).
Extended Data Figure 10 High-salt challenge in healthy human subjects.
a, Total salt intake according to dietary records (n = 12, paired one-tailed t-test). b, c, Metagenome analysis shows loss of Lactobacillus gut populations during human high-salt challenge. All subjects (x axis) for which gut Lactobacilli were detected are shown at baseline and all species detected (y axis) using the mOTU (b) or MetaPhlAn framework (c) for bacterial species identification are shown. Heat map cells show abundance of mOTU (insert counts as fraction of sample total) or average coverage (reads per position) for MetaPhlAn of the Lactobacillus species at baseline (left part of cells, black border) and after high-salt challenge (right part of cells, grey border). Cross markers show complete loss (no detection after high-salt challenge) of each species. In all cases but one (shown), baseline Lactobacillus populations are no longer detected after high salt intake. d, qPCR using Lactobacillus-specific 16S rDNA primers in human faecal samples positive for Lactobacillus at baseline show a loss of the respective species after 14 days of high salt. Lactobacillus 16S rDNA copy number in 4 ng faecal DNA is shown. Symbols indicate study subject, colours indicate respective Lactobacillus species. e, Kaplan–Meier survival curves contrasting the fate of gut Lactobacillus populations (detected using the mOTU framework) following a high-salt challenge (bright red curve) and in healthy control individuals from reference cohorts (n = 121, see Methods) not undergoing any intervention (bright blue curve). This is compared with corresponding survival curves over time for the set of all other detected gut bacterial species following high-salt challenge (high salt others, dark red curve) and without challenge in controls (NSD others, dark blue curve). f, For a clearer view of its time range, only the salt intervention curves from e are shown. Two observations are clear. First, Lactobacillus on average persist for shorter times in the gut than the average of all other species. Second, a high-salt challenge strongly increases gut loss of both Lactobacillus and non-Lactobacillus species. As such, in combination, Lactobacillus loss is highly pronounced after high-salt intervention and significantly (P < 1.62 × 10−8) faster than the average of all species. g–i, Metagenome analysis shows introduction of novel Lactobacillus gut populations during human high-salt challenge. All subjects (x axis) for which gut Lactobacilli were detected following high-salt challenge are shown, and all species detected (y axis) using the SpecI (g), mOTU (h) or MetaPhlAn (i) analysis are shown. Heat map cells show abundance (insert counts as fraction of sample total for SpecI and mOTU) and average coverage (reads per position for MetaPhlAn) of the Lactobacillus species at baseline (left part of cells, black border) and after high-salt challenge (right part of cells, grey border). Cross markers show novel introduction (no detection at baseline) of each species.
Supplementary information
Supplementary Information
This file contains Supplementary Results, including Supplementary Table 1 and Supplementary References. (PDF 171 kb)
Supplementary Data
This file contains metagenome analysis data. (XLS 69 kb)
Source data
Rights and permissions
About this article
Cite this article
Wilck, N., Matus, M., Kearney, S. et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551, 585–589 (2017). https://doi.org/10.1038/nature24628
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24628
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.