Abstract
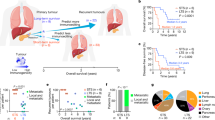
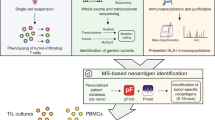
Pancreatic ductal adenocarcinoma is a lethal cancer with fewer than 7% of patients surviving past 5 years. T-cell immunity has been linked to the exceptional outcome of the few long-term survivors1,2, yet the relevant antigens remain unknown. Here we use genetic, immunohistochemical and transcriptional immunoprofiling, computational biophysics, and functional assays to identify T-cell antigens in long-term survivors of pancreatic cancer. Using whole-exome sequencing and in silico neoantigen prediction, we found that tumours with both the highest neoantigen number and the most abundant CD8+ T-cell infiltrates, but neither alone, stratified patients with the longest survival. Investigating the specific neoantigen qualities promoting T-cell activation in long-term survivors, we discovered that these individuals were enriched in neoantigen qualities defined by a fitness model, and neoantigens in the tumour antigen MUC16 (also known as CA125). A neoantigen quality fitness model conferring greater immunogenicity to neoantigens with differential presentation and homology to infectious disease-derived peptides identified long-term survivors in two independent datasets, whereas a neoantigen quantity model ascribing greater immunogenicity to increasing neoantigen number alone did not. We detected intratumoural and lasting circulating T-cell reactivity to both high-quality and MUC16 neoantigens in long-term survivors of pancreatic cancer, including clones with specificity to both high-quality neoantigens and predicted cross-reactive microbial epitopes, consistent with neoantigen molecular mimicry. Notably, we observed selective loss of high-quality and MUC16 neoantigenic clones on metastatic progression, suggesting neoantigen immunoediting. Our results identify neoantigens with unique qualities as T-cell targets in pancreatic ductal adenocarcinoma. More broadly, we identify neoantigen quality as a biomarker for immunogenic tumours that may guide the application of immunotherapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Ino, Y. et al. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br. J. Cancer 108, 914–923 (2013)
Hiraoka, N. et al. Intratumoral tertiary lymphoid organ is a favourable prognosticator in patients with pancreatic cancer. Br. J. Cancer 112, 1782–1790 (2015)
Remark, R. et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci. Immunol. 1, aaf6925 (2016)
Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013)
Gros, A. et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 22, 433–438 (2016)
Rizvi, N. A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128 (2015)
Hundal, J. et al. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med. 8, 11 (2016)
Witkiewicz, A. K. et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 6, 6744 (2015)
Zitvogel, L., Ayyoub, M., Routy, B. & Kroemer, G. Microbiome and anticancer immunosurveillance. Cell 165, 276–287 (2016)
Łuksza, M. & Lässig, M. A predictive fitness model for influenza. Nature 507, 57–61 (2014)
Verdegaal, E. M. E. et al. Neoantigen landscape dynamics during human melanoma–T cell interactions. Nature 536, 91–95 (2016)
Chekmasova, A. A. et al. Successful eradication of established peritoneal ovarian tumors in SCID-Beige mice following adoptive transfer of T cells genetically targeted to the MUC16 antigen. Clin. Cancer Res. 16, 3594–3606 (2010)
Morgado, M. et al. Tumor necrosis factor-α and interferon-γ stimulate MUC16 (CA125) expression in breast, endometrial and ovarian cancers through NFκB. Oncotarget 7, 14871–14884 (2016)
Das, S. et al. Carboxyl-terminal domain of MUC16 imparts tumorigenic and metastatic functions through nuclear translocation of JAK2 to pancreatic cancer cells. Oncotarget 6, 5772–5787 (2015)
Shukla, S. K. et al. MUC16-mediated activation of mTOR and c-Myc reprograms pancreatic cancer metabolism. Oncotarget 6, 19118–19131 (2015)
Muniyan, S. et al. MUC16 contributes to the metastasis of pancreatic ductal adenocarcinoma through focal adhesion mediated signaling mechanism. Genes Cancer 7, 110–124 (2016)
Elhanati, Y., Murugan, A., Callan, C. G., Mora, T. & Walczak, A. M. Quantifying selection in immune receptor repertoires. Proc. Natl Acad. Sci. USA 111, 9875–9880 (2014)
Saito, T. et al. Two FOXP3+CD4+ T cell subpopulations distinctly control the prognosis of colorectal cancers. Nat. Med. 22, 679–684 (2016)
Andersen, R. S. et al. Dissection of T-cell antigen specificity in human melanoma. Cancer Res. 72, 1642–1650 (2012)
Gross, L. Intradermal immunization of C3H mice against a sarcoma that originated in an animal of the same line. Cancer Res. 3, 326–333 (1943)
Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016)
Embuscado, E. E. et al. Immortalizing the complexity of cancer metastasis: genetic features of lethal metastatic pancreatic cancer obtained from rapid autopsy. Cancer Biol. Ther. 4, 548–554 (2005)
Haridas, D. et al. Pathobiological implications of MUC16 expression in pancreatic cancer. PLoS One 6, e26839 (2011)
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014)
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016)
Deshwar, A. G. et al. PhyloWGS: reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 16, 35 (2015)
Dharma Rao, T. et al. Novel monoclonal antibodies against the proximal (carboxy-terminal) portions of MUC16. Appl. Immunohistochem. Mol. Morphol. 18, 462–472 (2010)
Acknowledgements
We thank A. Rudensky, A. Snyder-Charan, C. Callan, Y. Elhanati, Z. Sethna, J. Leung, J. Ruan, C. Crabtree, P. Garcia, M. Singh, A. McNeil, D. Haviland, J. Melchor and J. Tsoi for discussions, technical and editorial assistance. This work was supported by National Institutes of Health (NIH) R01DK097087-01 Pancreatic Cancer Action Network-AACR Research Acceleration Network Grant (S.D.L.), P30 CA008748-50S4 administrative supplement (S.D.L., V.P.B.), Suzanne Cohn Simon Pancreatic Cancer Research Fund (S.D.L.), National Cancer Institute K12CA184746-01A1 (V.P.B.), Damon Runyon Clinical Investigator Award (V.P.B.), Stand Up to Cancer, Lustgarden Foundation, and the National Science Foundation (J.D.W., B.D.G.), the V Foundation (V.P.B., J.A.M., J.D.W., B.D.G.), the Phil A. Sharp Innovation Award (B.D.G., J.D.W.), Swim Across America, and the Ludwig Institute for Cancer Research (J.D.W., T.M.), and the Parker Institute for Cancer Immunotherapy (D.K.W., C.I.O.C., J.D.W., T.M.). Services by the Integrated Genomics Core were funded by the National Cancer Institute Cancer Center Support Grant (P30 CA08748), Cycle for Survival, and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Author information
Authors and Affiliations
Consortia
Contributions
V.P.B., M.Ł., P.J.A., D.T.F., J.D.W., R.P.D., B.D.G., T.A.C., T.M. and S.D.L. conceived the study and V.P.B., J.D.W., T.A.C., B.D.G., T.M. and S.D.L. designed all experiments. V.P.B., M.Ł., J.N.Z., V.M., J.A.M., R.R., B.H., G.A., U.B., Y.S., D.K.W., C.I.O.C., O.G.-H., M.A., B.M., J.Z., J.L., J.S., M.A.-A., R.Z., N.R., M.S., Z.L.K., O.B., A.J.L., P.J.A., D.T.F., M.M., S.G., C.A.I.-D. and members of the Australian Pancreatic Cancer Genome Initiative acquired and analysed data. G.A., U.B. and O.B. performed the histopathological analyses. M.A. and C.A.I.D. performed tissue acquisition, and mutational identification for rapid autopsy tissues. V.M., N.R., T.A.C., D.K.W. and C.I.O.C. performed the neoantigen identification. M.Ł. and B.D.G. constructed the neoantigen fitness models. V.P.B., J.N.Z., M.A.-A. and J.A.M. performed the in vitro T-cell assays. O.G.-H. performed the transfections, immunocytochemistry, and western blots. M.G. provided statistical oversight. V.P.B., M.Ł., J.N.Z., D.T.F., J.D.W., R.P.D., B.D.G., T.A.C., T.M. and S.D.L. interpreted the data. V.P.B., M.Ł., J.D.W., B.D.G., T.M. and S.D.L. drafted the manuscript.
Corresponding author
Ethics declarations
Competing interests
V.P.B., V.M., N.R., J.D.W. and T.A.C. have received research funding from Bristol-Myers Squibb. N.R. has received honoraria from MedImmune. D.T.F. is a co-founder of Myosotis LLC. S.G. has received research support from Immune Design, Janssen R&D, and Agenus, and serves on advisory boards for Third Rock, Ventures/Neon Therapeutics, B4CC, and Oncomed Pharmaceuticals. M.M., S.G. and R.R. are inventors of a patent regarding ‘Tissue profiling using multiplexed immunohistochemical consecutive staining’ (patent number pending). T.A.C. is a co-founder of Gritstone Oncology and is also an advisor for Genocea, Cancer Genetics, and Illumina.
Additional information
Reviewer Information Nature thanks E. Verdegaal, R. Vonderheide and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Clinicopathological characteristics of the MSKCC cohort.
a, Overall survival and patient overlap of short- and long-term survivors in tissue microarray, whole-exome sequencing, TCR Vβ sequencing, and bulk tumour transcriptomic profiling cohorts. b–g, Clinicopathological characteristics of patients in tissue microarray, transcriptome, TCR sequencing, whole-exome sequencing, matched primary–metastatic, and very long-term survivor cohorts. In b–e, the asterisk indicates three patients with metastases noted on final pathology (one liver metastasis, one metastasis to small bowel/mesentery, one splenic metastasis). n is the number of biologically independent samples in individual patients. P values were determined using a log-rank test (a) and a two-sided Fisher’s exact test (gender, tumour location, pN, pM, margin, chemotherapy), a two-sided χ2 test (procedure, pathological stage, pT, adjuvant treatment) and an unpaired, two-tailed Student’s t-test (age) (b–g).
Extended Data Figure 2 Long-term survivors of PDAC display enhanced intratumoural T-cell immunity.
a, Left: representative sequential immunohistochemical staining of a single short-term and a single long-term core tumour section. Sections bounded by black rectangles in the 100× panels are magnified to 275× in the panels immediately to the right for each core section. Top right: representative merged images of multiplexed immunohistochemistry are shown. The red rectangular sections are enlarged to 50×. CK19 was used to stain tumour cells. The arrows indicate CD3+CD8+granzyme B+ T cells. Middle right: immunofluorescent quantification of CD8+ and CD4+ cells in tumour tissue microarrays of short- and long-term survivors. Slides used were cut from separate sections of the block as those used for sequential immunohistochemistry (in a and Fig. 1b). Bottom right: quantification of multiplexed immunophenotyping as shown in a (left and top right). All immunofluorescent and immunohistochemical staining was repeated independently in triplicate for each patient. In a, short term n = 45, long term n = 51. b, Bulk tumour transcriptomic immune profiling in short- and long-term survivors. Dendritic-cell signature genes include CCL13, CCL17, CCL22, PPFIBP2, NPR1, HSD11B1 and CD209 (also known as DC-SIGN)3. c, Flow-cytometric gating strategy to phenotype human T cells (n = 7). The first plot is pre-gated on live cells, followed by CD45+ and CD3+CD56− cells. Values indicate the percentage of cells within the red boxes, and are gated based on isotype controls. d, Top: overall survival of patients who did or did not receive adjuvant chemotherapy (adjuvant chemotherapy+/− respectively, top left), and of patients with tumours harbouring more or fewer than the median number of CD3-CD8-granzyme B triple-positive cells (CD3-CD8-granzyme Bhi/low respectively, top right). Overall survival of all four groups is shown in the bottom panels. The table shows univariate and multivariate Cox regression analysis of clinicopathological features, adjuvant chemotherapy, and CD3-CD8-granzyme B density associations with overall survival. Horizontal bars indicate median values, error bars represent the s.e.m. n is the number of biologically independent samples in individual patients. P values were determined using a two-tailed Mann–Whitney U-test (a, b), a one-way ANOVA (c) and a log-rank test (d).
Extended Data Figure 3 Neoantigen quantity and CD8+ T-cell infiltrate identify long-term pancreatic cancer survivors.
a, Left: number of nonsynonymous, missense and neoantigenic mutations per patient in the MSKCC cohort. The tick marks on the x axis correspond to individual tumours. Right: oncoprint demonstrating the frequency of oncogenic driver-gene mutations in the MSKCC cohort. b, Overall survival of patients with tumours harbouring more than the median number of neoantigens (neoantigenhi), and greater than the median intratumoural T-cell repertoire polyclonality (polyclonalhi), compared to all other patients (rest). Neoantigens were determined using the MSKCC (top) and the pVAC-Seq (bottom) neoantigen prediction pipelines. c, Left: number of neoantigens per tumour, as determined by the MSKCC and pVAC-Seq neoantigen calling pipelines. Tick marks on the x axis correspond to individual tumours. Right: correlation matrix of neoantigens as determined by the MSKCC and pVAC-Seq neoantigen calling pipelines. The solid red line indicates the line of best fit, dotted lines indicate 95% confidence intervals. d, Top: overall survival of patients with tumours harbouring more or fewer than the median number of neoantigens (neoantigenhi/low) and CD3-CD8 double-positive cells (CD3-CD8hi/low), compared to all other patients (rest) (top left). Patients who did or did not receive adjuvant chemotherapy (adjuvant chemotherapy+/−, respectively) (top right), and all four groups (bottom) are also shown. The table shows univariate and multivariate Cox regression analysis of the associations of clinicopathological features, adjuvant chemotherapy, and neoantigen-CD3-CD8 number with overall survival. e Distribution of tumours with high- and low-quality neoantigens in neoantigenhi CD3-CD8hi long-term pancreatic cancer survivors compared to all other patients (rest). n is the number of biologically independent samples in individual patients. P values were determined using a log-rank test (b, d) and a χ2 test (e).
Extended Data Figure 4 Unique genomic features alone do not identify long-term survivors of PDAC.
a, Overall survival of patients with tumours harbouring more or fewer than the median number of neoantigens (neoantigenhi/low), CD3-CD8 double-positive cells (CD3-CD8hi/low), polyclonality (polyclonalhi/low), mutations (mutationhi/low), and CD4 single positive cells (CD4hi/low). b, Oncoprint demonstrating no difference in the frequency of oncogenic driver mutations in short- and long-term tumours. c, No difference was found in the number of nonsynonymous, missense, and immunogenic mutations (neoantigens) in short- and long-term PDAC tumours. d, Overall survival stratified by mutations in ARID1A, KRASQ61H, RBM10 and MLL-related genes (KMT2A, KMT2B, KMT2C and KMT2E (also known as MLL, MLL2, MLL3 and MLL5, respectively)). Horizontal bars indicate median values. n is the number of biologically independent samples in individual patients. P values were determined using a log-rank test (a, d).
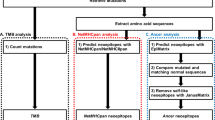
Extended Data Figure 5 Neoantigen immune fitness models.
a, Comprehensive flowchart of neoantigen quality identification pipeline. Software programs used for each step are indicated in italicized text. Mathematical formulae for the calculation of individual components of neoantigen quality are defined in the Methods. All software components of the pipeline are published and/or publicly available. b, Top: schematic of neoantigen immune fitness models. Each circle represents a tumour clone in an evolutionary tree. Clones in both models are identical with respect to the number of mutations and neoantigens. The numbers represent hypothetical neoantigens gained in a successive tumour clone. Shades of red indicate the immunogenicity of each clone, as ascribed by the two models, namely neoantigen quality or neoantigen quantity. Bottom: parameters defining the quality score in the quality model (1)–(3). In (1), amino acid sequences of a hypothetical wild-type epitope, tumour neoepitope, and a homologous microbial epitope are shown. Yellow highlights the changing amino acid between the wild-type and the tumour sequence as a consequence of a tumour-specific mutation. The amino acids in red indicate homology between the tumour neoepitope and the microbial epitope.
Extended Data Figure 6 Neoantigen quality is independently prognostic of survival.
a, Top: overall survival of patients whose tumours displayed high compared to low neoantigen quality (neoantigen qualityhi/low) (left), and overall survival of patients who did or did not receive adjuvant chemotherapy (right). Bottom: overall survival of all four groups. Neoantigen quality defined by pipeline and schema as defined in Extended Data Fig. 5a, b. The table shows univariate and multivariate Cox regression analyses of the associations of clinicopathological features, adjuvant chemotherapy and neoantigen quality with overall survival. Data include all patients in the whole-exome sequencing MSKCC cohort. b, Number of nonsynonymous, missense and neoantigenic mutations per patient in the ICGC cohort (n = 166). c, Top: overall survival of patients in the ICGC cohort whose tumours displayed high compared to low neoantigen quality (neoantigen qualityhi/low) (left), and overall survival of patients in the ICGC cohort stratified by adjuvant chemotherapy administration (right). Bottom: overall survival of all four groups. Neoantigen quality defined by pipeline and schema as defined in Extended Data Fig. 5a, b. The table shows univariate and multivariate Cox regression analyses of the associations of clinicopathological features, adjuvant chemotherapy and neoantigen quality with overall survival in the ICGC cohort. Data on adjuvant chemotherapy is included for patients whose treatment status was available. n is the number of biologically independent samples in individual patients. P values were determined using a log-rank test (a, c).
Extended Data Figure 7 Stability of neoantigen quality model parameters on subsampled cohorts and prognostic dependence of neoantigen quality on infectious disease-derived peptides.
a, b, Parameters of the neoantigen fitness quality model for the MSKCC (a) and the ICGC (b) cohorts. Left: log-rank test score landscape as a function of the model parameters (the horizontal alignment score displacement a, and the characteristic time τ); the significance of the score is denoted in the legend. Right: two-dimensional histograms showing distributions of optimal parameters obtained on subsampled datasets with 50, 70, and 90% of patients left, over 500 iterations of subsampling at each frequency. c, Overall survival of patients in the MSKCC and ICGC cohorts; whole tumours displayed high compared to low neoantigen quality (neoantigen qualityhi/low). Neoantigen quality was calculated using alignment to immunogenic infectious disease-derived IEDB peptides (microbial peptides) or using alignment to immunogenic non-infectious disease-derived allergy or autoimmune peptides in the IEDB database (non-microbial peptides). n is the number of biologically independent samples in individual patients. P values were determined using a log-rank test (c).
Extended Data Figure 8 Predicted MUC16 neoantigens are recognized by the human TCR repertoire.
a, PBMCs pulsed with no peptide, wild-type control peptide, cross-reactive peptide or high-quality neopeptide (mutant). Representative gating strategies for CD8+ T-cell expansion and degranulation are shown. b, PBMCs pulsed with no peptide, wild-type control peptide, and MUC16 neopeptides (mutant). Representative gating strategies for CD8+ T-cell expansion are shown. c, Representative gating strategy to identify CD8+ T cells in peripheral blood of healthy donors (top panel). Identification of CD8+ T cells in healthy donors reactive to unique MUC16 neoepitopes predicted to bind to the B*0801 HLA-allele, using MUC16-neoepitope–HLA multimers. Quantification of all healthy donors (neoepitope no. 1, 2, n = 5) is shown (right). Multimer staining is shown on the x axis, CD8 is shown on the y axis. Peptide information is provided in Supplementary Table 3. n is the number of biologically independent samples in individual patients. Horizontal bars indicate median values, error bars represent the s.e.m. P values were determined using a one-way ANOVA (c).
Extended Data Figure 9 Long-term survivors do not display differences in MUC16 mutations, transcriptional regulators or downstream targets of MUC16, or differences in other mucins and tissue expression antigens.
a, The frequency of MUC16 mutations in short- and long-term PDAC tumours. Lollipop plot showing the location of MUC16 mutations and neoantigens in short- and long-term survivors of pancreatic cancer. b, Expression of bulk tumour MUC16 mRNA (left) and protein (middle) by immunohistochemistry. Immunohistochemical staining was repeated independently in triplicate for each patient. Right: MUC16 mutant allele frequency in non-hypermutated tumours with MUC16 mutations. c, Left: frequency of patients with MUC16 neoantigens in MSKCC and ICGC cohorts. Middle: frequency of patients with neoantigens in genes recurrently harbouring neoantigens in >5% of patients in both MSKCC and ICGC cohorts. Right: genes most frequently harbouring neoantigens in the MSKCC cohort as determined by pVAC-Seq. Frequency of patients (y axis) and raw numbers (above bar graphs) are indicated. d, mRNA expression of transcriptional activators of MUC16 (top left), mediators implicated in MUC16-dependent tumour progression (top right), and mRNA (bottom left) and protein (bottom right) levels of tissue expression antigens MUC1, MUC4, WT1, mesothelin and annexin A2 in short- and long-term tumours. WT1 protein was undetectable in both short- and long-term survivors. n = 15 per group in top left, top right, and bottom left; short term n = 45, long term n = 51 in bottom right. e, MUC16 mRNA and protein expression in MUC16 non-mutated (WT; n = 18 (top), n = 20 (bottom)) and mutated (mutant; n = 10 (top), n = 9 (bottom)) tumours. f, TCR Vβ sequencing of T-cell product following the pulse of peripheral blood T cells with MUC16 neopeptides as in Fig. 4e. Brown open circles indicate stable or contracted clones with mutant neopeptide; blue open circles indicate expanded clones with mutant neopeptide; red solid circles indicate expanded clones with mutant neopeptide detected in archival primary tumours. Arrows indicate clones in archival primary tumours with rank frequencies; Venn diagrams show clonal overlap in respective compartments. Horizontal bars indicate median values, error bars represent the s.e.m. n is the number of biologically independent samples in individual patients. P values were determined using two-tailed Mann–Whitney U- and Student’s t-tests (b), a χ2 test (c) and as described in the Methods (f).
Extended Data Figure 10 MUC16 mutations do not alter tumour cell-intrinsic MUC16 protein expression.
a, Representative immunohistochemical staining (left) and quantification (right) of MUC16 expression in tissue microarrays of short- and long-term survivors of pancreatic cancer, as assessed using three independent anti-MUC16 antibodies. Ab no. 1: clone EPSISR23, purchased from Abcam; Ab no. 2: polyclonal, purchased from Abcam ab133419; Ab no. 3: clone 4H1127. Each open circle represents the median expression of independent immunohistochemical staining performed in triplicate for each patient. b, Western blot (top) and immunocytochemistry (bottom) of untransfected (−), empty vector (vector), MUC16 wild-type (MUC16 WT) and MUC16 mutant (MUC16 R15C) HEK293 cells. The left blot was probed with anti-MUC16-specific antibody (clone 4H1127) and the right blot with anti-β-actin. The red rectangle indicates the MUC16-specific band. All cells in the bottom panels were probed with anti-MUC16 antibody (clone 4H1127). The inserted mutation was identical to a neoantigenic MUC16 mutation (detected in patient 1 shown in Extended Data Fig. 9a). Data are representative of two independent experiments with similar results. c, MUC16 immunohistochemistry on samples from two long-term survivors of pancreatic cancer with MUC16 neoepitopes in primary resected tumours. Areas in rectangular low-power fields are shown magnified in the right panels. Immunohistochemical staining was performed independently in triplicate for each patient in tissue microarrays, and confirmed with immunohistochemical staining on whole tumour sections (shown). Horizontal bars indicate median values, error bars represent the s.e.m. n is the number of biologically independent samples in individual patients. P values were determined using a two-tailed Student’s t-test (a).
Supplementary information
Supplementary Table 1
This file contains a comprehensive list of all neopeptide sequences and HLA alleles with predicted neopeptide binding in the MSKCC cohort. (XLSX 334 kb)
Supplementary Table 2
This file contains a comprehensive list of all human infectious derived, class-I restricted peptide sequences with positive immune assays derived from the Immune Epitope Database used in this study. (XLSX 110 kb)
Supplementary Table 3
This file contains a comprehensive list of all peptide sequences, respective HLA alleles with predicted binding, crossreacting microbial sequences, and microbial species (where applicable) used in this study. (XLSX 40 kb)
Supplementary Table 4
This file contains a comprehensive list of all human non-infectious (allergy/autoimmune) derived, class-I restricted peptide sequences with positive immune assays derived from the Immune Epitope Database used in this study. (XLSX 53 kb)
Supplementary Data 1
This folder contains the source code for the neoantigen quality assessment performed in this manuscript. A text file of Supplementary Table 1 is included to enable code execution. (ZIP 764 kb)
Source data
Rights and permissions
About this article
Cite this article
Balachandran, V., Łuksza, M., Zhao, J. et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017). https://doi.org/10.1038/nature24462
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature24462
This article is cited by
-
Characterizing and forecasting neoantigens-resulting from MUC mutations in COAD
Journal of Translational Medicine (2024)
-
Genetically engineered membrane-based nanoengagers for immunotherapy of pancreatic cancer
Journal of Nanobiotechnology (2024)
-
Novel insights into TCR-T cell therapy in solid neoplasms: optimizing adoptive immunotherapy
Experimental Hematology & Oncology (2024)
-
Therapeutic developments in pancreatic cancer
Nature Reviews Gastroenterology & Hepatology (2024)
-
Computational immunogenomic approaches to predict response to cancer immunotherapies
Nature Reviews Clinical Oncology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.