Abstract
P53-binding protein 1 (53BP1) is a multi-functional double-strand break repair protein that is essential for class switch recombination in B lymphocytes and for sensitizing BRCA1-deficient tumours to poly-ADP-ribose polymerase-1 (PARP) inhibitors. Central to all 53BP1 activities is its recruitment to double-strand breaks via the interaction of the tandem Tudor domain with dimethylated lysine 20 of histone H4 (H4K20me2). Here we identify an uncharacterized protein, Tudor interacting repair regulator (TIRR), that directly binds the tandem Tudor domain and masks its H4K20me2 binding motif. Upon DNA damage, the protein kinase ataxia-telangiectasia mutated (ATM) phosphorylates 53BP1 and recruits RAP1-interacting factor 1 (RIF1) to dissociate the 53BP1–TIRR complex. However, overexpression of TIRR impedes 53BP1 function by blocking its localization to double-strand breaks. Depletion of TIRR destabilizes 53BP1 in the nuclear-soluble fraction and alters the double-strand break-induced protein complex centring 53BP1. These findings identify TIRR as a new factor that influences double-strand break repair using a unique mechanism of masking the histone methyl-lysine binding function of 53BP1.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Panier, S. & Boulton, S. J. Double-strand break repair: 53BP1 comes into focus. Nature Rev. Mol. Cell Biol. 15, 7–18 (2014)
Zimmermann, M. & de Lange, T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 24, 108–117 (2014)
Bothmer, A. et al. Mechanism of DNA resection during intrachromosomal recombination and immunoglobulin class switching. J. Exp. Med. 210, 115–123 (2011)
Dong, J. et al. Orientation-specific joining of AID-initiated DNA breaks promotes antibody class switching. Nature 525, 134–139 (2015)
Ward, I. M. et al. 53BP1 is required for class switch recombination. J. Cell Biol. 165, 459–464 (2004)
Manis, J. P. et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nature Immunol. 5, 481–487 (2004)
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005)
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005)
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010)
Bouwman, P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature Struct. Mol. Biol. 17, 688–695 (2010)
Chapman, J. R., Sossick, A. J., Boulton, S. J. & Jackson, S. P. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci. 125, 3529–3534 (2012)
Charier, G. et al. The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure 12, 1551–1562 (2004)
Iwabuchi, K. et al. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J. Biol. Chem. 278, 36487–36495 (2003)
Fradet-Turcotte, A. et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 (2013)
Botuyan, M. V. et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 (2006)
Jowsey, P. et al. Characterisation of the sites of DNA damage-induced 53BP1 phosphorylation catalysed by ATM and ATR. DNA Repair 6, 1536–1544 (2007)
Ward, I. M., Minn, K., Jorda, K. G. & Chen, J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 278, 19579–19582 (2003)
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009)
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009)
Baciu, P. C. et al. Syndesmos, a protein that interacts with the cytoplasmic domain of syndecan-4, mediates cell spreading and actin cytoskeletal organization. J. Cell Sci. 113, 315–324 (2000)
Denhez, F. et al. Syndesmos, a syndecan-4 cytoplasmic domain interactor, binds to the focal adhesion adaptor proteins paxillin and Hic-5. J. Biol. Chem. 277, 12270–12274 (2002)
Taylor, M. J. & Peculis, B. A. Evolutionary conservation supports ancient origin for Nudt16, a nuclear-localized, RNA-binding, RNA-decapping enzyme. Nucleic Acids Res. 36, 6021–6034 (2008)
Kim, H. et al. Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan. Biochem. Biophys. Res. Commun. 463, 762–767 (2015)
Lemaître, C. et al. The nucleoporin 153, a novel factor in double-strand break repair and DNA damage response. Oncogene 31, 4803–4809 (2012)
Orthwein, A. et al. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science 344, 189–193 (2014)
Lee, D. H. et al. Dephosphorylation enables the recruitment of 53BP1 to double-strand DNA breaks. Mol. Cell 54, 512–525 (2014)
Hartlerode, A. J. et al. Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks. PLoS ONE 7, e49211 (2012)
Mertins, P. et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nature Methods 10, 634–637 (2013)
Chapman, J. R. et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49, 858–871 (2013)
Di Virgilio, M. et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science 339, 711–715 (2013)
Escribano-Diaz, C. et al. A cell cycle-dependent regulatory circuit composed of 53BP1–RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 (2013)
Zimmermann, M., Lottersberger, F., Buonomo, S. B., Sfeir, A. & de Lange, T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 339, 700–704 (2013)
Feng, L., Fong, K. W., Wang, J., Wang, W. & Chen, J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J. Biol. Chem. 288, 11135–11143 (2013)
Callen, E. et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 153, 1266–1280 (2013)
Zong, D. et al. Ectopic expression of RNF168 and 53BP1 increases mutagenic but not physiological non-homologous end joining. Nucleic Acids Res. 43, 4950–4961 (2015)
Acs, K. et al. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nature Struct. Mol. Biol. 18, 1345–1350 (2011)
Mallette, F. A. et al. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31, 1865–1878 (2012)
Tang, J. et al. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nature Struct. Mol. Biol. 20, 317–325 (2013)
Gudjonsson, T. et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 150, 697–709 (2012)
Lu, R. & Wang, G. G. Tudor: a versatile family of histone methylation ‘readers’. Trends Biochem. Sci. 38, 546–555 (2013)
Wagner, T., Robaa, D., Sippl, W. & Jung, M. Mind the methyl: methyllysine binding proteins in epigenetic regulation. ChemMedChem 9, 466–483 (2014)
Sandhu, S. K., Yap, T. A. & de Bono, J. S. The emerging role of poly(ADP-Ribose) polymerase inhibitors in cancer treatment. Curr. Drug Targets 12, 2034–2044 (2011)
Yap, T. A., Sandhu, S. K., Carden, C. P. & de Bono, J. S. Poly(ADP-ribose) polymerase (PARP) inhibitors: exploiting a synthetic lethal strategy in the clinic. CA Cancer J. Clin. 61, 31–49 (2011)
Lord, C. J. & Ashworth, A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nature Med. 19, 1381–1388 (2013)
Drané, P., Ouararhni, K., Depaux, A., Shuaib, M. & Hamiche, A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24, 1253–1265 (2010)
Macnaughtan, M. A., Kane, A. M. & Prestegard, J. H. Mass spectrometry assisted assignment of NMR resonances in reductively 13C-methylated proteins. J. Am. Chem. Soc. 127, 17626–17627 (2005)
Cui, G., Botuyan, M. V. & Mer, G. Preparation of recombinant peptides with site- and degree-specific lysine 13C-methylation. Biochemistry 48, 3798–3800 (2009)
Simon, M. D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007)
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995)
Johnson, B. A. & Blevins, R. A. NMR View: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994)
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010)
Acknowledgements
We are grateful to L. Xu for RIF1 antibody and S. Boulton for RIF1−/− MEFs. D.C. is supported by R01 AI101897-01 (National Institute of Allergy and Infectious Diseases) and R01CA142698-07 (National Cancer Institute), a Leukemia and Lymphoma Society Scholar Grant, the Claudia Adams Barr Program for Innovative Cancer Research, a Department of Defense Ovarian Cancer Award, a Breast SPORE Pilot Award, and a Robert and Deborah First Fund Award. G.M. is supported by National Institutes of Health grants R01 CA132878 and R01 GM116829, and a Mayo Clinic Brain Cancer SPORE Program Pilot Award (P50 CA108961). J.W.H. is supported by AG011085. J.C. is supported by National Institutes of Health/National Institute of Allergy and Infectious Disease grants (1RO1AI072194 and 1RO1AI124186) and a National Cancer Institute Cancer Center Support grant (P30CA008748). W.T.Y. is supported by a training grant from the National Cancer Institute (4T32CA009149-40).
Author information
Authors and Affiliations
Contributions
P.D., G.M. and D.C. designed the study. P.D. performed most of the experiments with assistance from M.E.B., K.M., S.C., Y.Z.H., X.F.F. and N.P. A.D. did the statistical analysis. G.C. and M.V.B. conducted NMR studies under G.M.’s supervision. A.K. conducted microscopy studies with the LacO fusion system under E.S.’s guidance. C.M. did quantitative MS under J.W.H.’s guidance. W.T.Y. did CSR assays under J.C.’s guidance. P.D., G.M. and D.C. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks D. Durocher, J. Jonkers and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
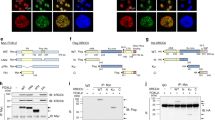
Extended Data Figure 1 TIRR prevents the interaction of 53BP1 Tudor domain with a peptide derived from H4K20me2.
a, Soluble nuclear proteins were prepared from HeLa and HeLa cells in which TIRR has been knock out using CRISPR/cas9 system (HeLaΔTIRR) undamaged or irradiated at 10 Gy for 90 min. Endogenous 53BP1 was next immunoprecipitated and immunoblotted using the indicated antibody. b, Coomassie staining of Flag-tagged recombinant proteins purified from bacteria; ‘OD-Tudor’ encompasses the oligomerization, the Tudor and UDR domains, ‘OD’ the oligomerization domain and ‘Tudor-UDR’ the Tudor and UDR domains. c, Recombinant TIRR-His protein was incubated with the indicated Flag fusion protein in presence of 200 or 400 mM NaCl. Pull-down proteins were subjected to Flag (top) and His (bottom) immunoblotting. M, molecular size markers. d, Coomassie staining of Flag–OD–Tudor and Flag–OD–Tudor/TIRR–His complex purified from bacteria co-expressing both proteins. e, Pull-down of the indicated proteins with a biotinylated peptide derived from histone H4K20me2. Bound proteins were analysed by immunoblotting. f, Interaction of 53BP1 Tudor with TIRR probed using ITC. Integrated heat measurements from raw titration data and curve fitting with a standard one-site model are shown. Kd and stoichiometry (n) are indicated.
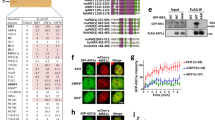
Extended Data Figure 2 Ectopic TIRR restores RAD51 foci formation in BRCA1-mutant MEFs.
a, Brca1-mutant MEFs stably transduced with an empty vector (EV) or TIRR-FH were transfected with the indicated siRNA. RAD51 was stained 6 h after 5 Gy irradiation (mean ± s.d., n = 3). b, Quantification of CSR at 96 h or 72 h after stimulation from splenic B cells transduced with pMIG or pMIG–TIRR, and stimulated with LPS and IL-4 for 72 h. c, AID, Sμ germline (Sμ GLT) and Sγ1 germline (Sγ1 GLT, black) transcripts in splenic B cells stimulated for 48 h (mean ± s.d., n = 3 mice). d, Immunoblotting of AID and TIRR from lysates prepared from splenic B cells stimulated for 72 h.
Extended Data Figure 3 TIRR expression level remains unchanged after irradiation.
a, TIRR-FH-expressing U2OS cells were irradiated at 10 Gy and collected next at the indicated time. Whole-cell extracts were analysed by immunoblotting using the indicated antibody. b, U2OS whole-cell extracts were prepared 90 min after a 10 Gy irradiation and analysed by immunoblotting using the indicated antibody. c, RPE-1 cells stably expressing TIRR-FH wild-type (WT) or double mutated (K10R, K151R) were irradiated at 10 Gy for 90 min followed by TIRR-FH immunoprecipitation. Pull-down proteins were subjected to immunoblotting with the indicated antibodies. d, U2OS cells were first transfected with the indicated siRNA. The day after, cells were transfected with a vector encoding Ubiquitin fused to an N-terminal His tag. Forty-eight hours later, cells were irradiated at 10 Gy for 90 min when indicated followed by His pull-down. Pull-down proteins were subjected to immunoblotting with the indicated antibodies. e, Laser stripes examined by immunofluorescence 15 min after irradiation in RPE-1 cells expressing TIRR-FH (left) or RPE-1 cells (right).
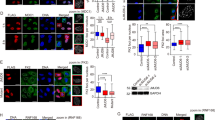
Extended Data Figure 4 RIF1 depletion affects 53BP1 foci formation in various cellular models.
a, RPE-1 cells deprived of endogenous ATM using CRISPR/cas9 system (RPE-1 ΔATM) were stably transfected with GFP–TIRR. GFP–TIRR was next pull-down and retained proteins were analysed by immunoblotting. The star represents a non-specific band. b, Flag immunoprecipitation from extracts prepared from indicated siRNA-transfected TIRR-FH-expressing RPE-1 cells. c, Immunofluorescence of 53BP1 and RIF1 in siRNA-transfected U2OS cells irradiated at 10 Gy for 90 min. The graph represents the percentage of cells harbouring more than ten 53BP1 foci. d, Same as c in RPE-1 cells (mean ± s.d., n = 2). e, Kinetics of 53BP1 foci formation in RPE-1 ΔRIF1 cells. Results are expressed as number of foci per cell (top) and as percentage of cells harbouring more than ten 53BP1 foci (bottom) (mean ± s.d., n = 2). Deletion of RIF1 was checked by immunoblotting. f, Same as in e in MEF RIF1−/− cells.
Extended Data Figure 5 TIRR depletion affects mainly the stability of soluble nuclear 53BP1.
a, Immunoblotting of siRNA-transfected Hela cell extracts. TIRR expression has been rescued with an siRNA-resistant construct. b, Nuclear proteins were sequentially extracted by increasing amount of salt from siRNA-transfected U2OS cells. After salt extraction, the resulting pellet corresponds to the insoluble material (insoluble). The amount of 53BP1 present in each fraction was measured by immunoblotting and quantification using ImageJ. c, Relative TIRR and 53BP1 protein distribution in the RPE-1 nucleus as measured by mass spectrometry. d, U2OS-soluble nuclear extract (1 mg) was loaded onto a sucrose gradient of 5–30% and ultracentrifuged for 4 h at 40,000 rpm. Fractions were collected from the top of the gradient and immunoblotted. The amount of 53BP1 and TIRR was quantified using ImageJ. e, Quantification of TIRR mRNA expression in siRNA-transfected Brca1-mutant MEFs. TIRR level was normalized to 5S expression.
Extended Data Figure 6 Expression of TIRR protein in various cancer cell lines and alteration frequency of Nudt16L1 (TIRR) gene in cancer.
a, Immunoblotting of 53BP1 and TIRR from extracts prepared from various human cancer cell lines: A, Ovarian cancer cell lines; B, Breast cancer cell lines; C, paediatric glioma cell lines; D, pancreatic cell lines; E, lung cell lines. HCT116, colon cancer cells; Jurkat, acute T cell leukaemia; LnCap, prostate adenocarninoma. Epithelial cells from retinal pigment RPE-1 and ovarian surface HiO80 are immortalized cell lines. b, Data extracted from cBioportal (http://www.cBioportal.org) for alterations in the Nudt16L1 (TIRR) gene across different cancer types. Twenty-nine out of 34 cancer types exhibit amplifications in the Nudt16L1 gene.
Extended Data Figure 7 TIRR depletion modifies the interaction of 53BP1 with its partners.
a, HeLa cells deprived of endogenous TIRR (HeLaΔTIRR) were stably transduced with FH-53BP1 together with GFP–TIRR or GFP. FH-53BP1 partners were purified from total nuclear extracts (nuclear-soluble and chromatin extracts) 90 min after a 10 Gy irradiation and analysed by mass spectrometry. b, HeLaΔTIRR or parental HeLa cells were stably transduced with FH-53BP1. FH-53BP1 partners were purified from total nuclear extracts from undamaged or irradiated cells and analysed by immunoblotting after normalization of the amount of 53BP1 pull-down (see results). The star represents a non-specific band.
Extended Data Figure 8 TIRR depletion hypersensitizes human BRCA1-mutant cells to PARPi in a 53BP1-dependent manner.
Human ovarian cancer cell lines Cov362 (a) and UWB1.289 (b) were transfected with the indicated siRNA, followed by treatment with olaparib. Percentage survival was calculated by normalizing the survival from treatment with olaparib versus untreated cells (mean ± s.d., n = 3). Efficiency of siRNAs was checked by immunoblot using the indicated antibodies in the Cov362 cell line. c, Top: siRNA-transfected Brca1-mutant MEFs mock-transfected or overexpressing RNF168 were treated with olaparib. Percentage survival was calculated as in a and b (mean ± s.d., n = 3). Bottom: stable expression of Flag–RNF168 and efficiency of 53BP1 siRNA were checked by immunoblotting. d, Immunofluorescence of 53BP1 in BRCA1-mutant MEFs (mean ± s.d., n = 2).
Extended Data Figure 9 TIRR depletion increases radiosensitivity.
a, Whole-cell extracts from RPE-1 cells (CTRL) and from a polyclonal population (poly) and three RPE-1 clones deprived of endogenous TIRR using CRISPR/cas9 system were analysed by immunoblotting using the indicated antibodies. The amount of 53BP1 was quantified and normalized to β-tubulin using ImageJ. b, Kinetics of γH2AX foci formation in indicated cells after a 2 Gy irradiation. The graph represents the mean number of γH2AX foci per cell (mean ± s.d., n = 3). c, Clonogenic survival of indicated cells after irradiations (0 to 5 Gy). TIRR expression has been restored in clone 13 by stable expression of GFP–TIRR. Survival was expressed as a percentage of colonies formed relative to the non-irradiated control (mean ± s.d., n = 3).
Supplementary information
Supplementary Figure 1
This file contains the uncropped scans for Immunoblot and silver stained gels with protein sizes indicated in kDa. (PDF 3654 kb)
Supplementary Table 1
This file contains the mass spectrometry analyses of FH-53BP1-FFR complex purified from U2OS cells. (XLSX 51 kb)
Supplementary Table 2
This file contains the mass spectrometry analyses of TIRR-FH complex purified from U2OS cells. (XLSX 82 kb)
Supplementary Table 3
This file contains the mass spectrometry analyses of FH-53BP1 complex purified from HeLa△TIRR stably expressing GFP-TIRR or GFP. Cells were irradiated at 10 Gy for 90 min before purification of 53BP1-complex by tandem affinity. (XLSX 80 kb)
Rights and permissions
About this article
Cite this article
Drané, P., Brault, ME., Cui, G. et al. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature 543, 211–216 (2017). https://doi.org/10.1038/nature21358
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21358
This article is cited by
-
An autoinhibited state of 53BP1 revealed by small molecule antagonists and protein engineering
Nature Communications (2023)
-
TRABID overexpression enables synthetic lethality to PARP inhibitor via prolonging 53BP1 retention at double-strand breaks
Nature Communications (2023)
-
Dynamics of the DYNLL1–MRE11 complex regulate DNA end resection and recruitment of Shieldin to DSBs
Nature Structural & Molecular Biology (2023)
-
Gene losses may contribute to subterranean adaptations in naked mole-rat and blind mole-rat
BMC Biology (2022)
-
Identification of a novel GR-ARID1a-P53BP1 protein complex involved in DNA damage repair and cell cycle regulation
Oncogene (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.