Abstract
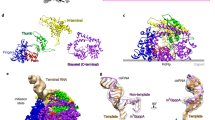
Viruses in the Reoviridae, like the triple-shelled human rotavirus and the single-shelled insect cytoplasmic polyhedrosis virus (CPV), all package a genome of segmented double-stranded RNAs (dsRNAs) inside the viral capsid and carry out endogenous messenger RNA synthesis through a transcriptional enzyme complex (TEC)1. By direct electron-counting cryoelectron microscopy and asymmetric reconstruction, we have determined the organization of the dsRNA genome inside quiescent CPV (q-CPV) and the in situ atomic structures of TEC within CPV in both quiescent and transcribing (t-CPV) states. We show that the ten segmented dsRNAs in CPV are organized with ten TECs in a specific, non-symmetric manner, with each dsRNA segment attached directly to a TEC. The TEC consists of two extensively interacting subunits: an RNA-dependent RNA polymerase (RdRP) and an NTPase VP4. We find that the bracelet domain of RdRP undergoes marked conformational change when q-CPV is converted to t-CPV, leading to formation of the RNA template entry channel and access to the polymerase active site. An amino-terminal helix from each of two subunits of the capsid shell protein (CSP) interacts with VP4 and RdRP. These findings establish the link between sensing of environmental cues by the external proteins and activation of endogenous RNA transcription by the TEC inside the virus.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Electron Microscopy Data Bank
Protein Data Bank
Data deposits
3D cryo-electron microscopy (cryoEM) density maps have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-6408 (3.3 Å averaged TEC in q-CPV), EMD-6404 (4.0 Å averaged TEC in t-CPV), EMD-6407 (3.9 Å full q-CPV), EMD-6405 (4.8 Å full t-CPV), EMD-6406 (5.1 Å asymmetric reconstruction of q-CPV by capsid-subtraction method) and EMD-6409 (filtered 22 Å q-CPV asymmetric reconstruction). The coordinates of atomic models of the TEC in q-CPV and t-CPV have been deposited in the Protein Data Bank under accession number 3JB6 and 3JB7, respectively.
Change history
30 October 2015
Affiliations were amended in the HTML on 30 October 2015
25 November 2015
A minor change was made to the labelling in Fig. 2b, and ‘Arg979’ was corrected to ‘Arg997’ in the main text.
References
Mertens, P. P. C., Rao, S. & Zhou, Z. H. in Virus Taxonomy, VIIIth Report of the ICTV (eds Fauquet, C. M. et al.) 522–533 (Elsevier/Academic Press, 2004)
Tao, Y., Farsetta, D. L., Nibert, M. L. & Harrison, S. C. RNA synthesis in a cage—structural studies of reovirus polymerase λ3. Cell 111, 733–745 (2002)
Lu, X. et al. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure 16, 1678–1688 (2008)
Jiang, W. et al. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439, 612–616 (2006)
Lander, G. C. et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312, 1791–1795 (2006)
Zhou, Z. H. in Segmented Double-Stranded RNA Viruses: Structure and Molecular Biology (ed. Patton, J. T. ) 27–43 (Caister Academic Press, 2008)
Furuichi, Y . “Methylation-coupled” transcription by virus-associated transcriptase of cytoplasmic polyhedrosis virus containing double-stranded RNA. Nucleic Acids Res. 1, 809–822 (1974)
Estrozi, L. F. et al. Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles. J. Mol. Biol. 425, 124–132 (2013)
Zhang, X., Walker, S. B., Chipman, P. R., Nibert, M. L. & Baker, T. S. Reovirus polymerase λ3 localized by cryo-electron microscopy of virions at a resolution of 7.6Å. Nature Struct. Mol. Biol. 10, 1011–1018 (2003)
Nason, E. L. et al. Interactions between the inner and outer capsids of bluetongue virus. J. Virol. 78, 8059–8067 (2004)
Xia, Q., Jakana, J., Zhang, J.-Q. & Zhou, Z. H. Structural comparisons of empty and full cytoplasmic polyhedrosis virus protein-RNA interactions and implications for endogenous RNA transcription mechanism. J. Biol. Chem. 278, 1094–1100 (2003)
Gouet, P. et al. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 97, 481–490 (1999)
Abels, J., Moreno-Herrero, F., Van der Heijden, T., Dekker, C. & Dekker, N. Single-molecule measurements of the persistence length of double-stranded RNA. Biophys. J. 88, 2737–2744 (2005)
Nibert, M. L. & Kim, J. Conserved sequence motifs for nucleoside triphosphate binding unique to turreted Reoviridae members and coltiviruses. J. Virol. 78, 5528–5530 (2004)
Zhao, S., Liang, C., Hong, J. & Peng, H. Genomic sequence analyses of segments 1 to 6 of Dendrolimus punctatus cytoplasmic polyhedrosis virus. Arch. Virol. 148, 1357–1368 (2003)
Sutton, G., Grimes, J. M., Stuart, D. I. & Roy, P. Bluetongue virus VP4 is an RNA-capping assembly line. Nature Struct. Mol. Biol. 14, 449–451 (2007)
Stauber, N., Martinez-Costas, J., Sutton, G., Monastyrskaya, K. & Roy, P. Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J. Virol. 71, 7220–7226 (1997)
Kim, J., Parker, J. S., Murray, K. E. & Nibert, M. L. Nucleoside and RNA triphosphatase activities of orthoreovirus transcriptase cofactor μ2. J. Biol. Chem. 279, 4394–4403 (2004)
Choi, K. H. & Rossmann, M. G. RNA-dependent RNA polymerases from Flaviviridae. Curr. Opin. Struct. Biol. 19, 746–751 (2009)
Yang, C. et al. Cryo-EM structure of a transcribing cypovirus. Proc. Natl Acad. Sci. USA 109, 6118–6123 (2012)
Yu, X., Jiang, J., Sun, J. & Zhou, Z. H. A. putative ATPase mediates RNA transcription and capping in a dsRNA virus. Elife 4, e07901 (2015)
Luongo, C. L. et al. Loss of activities for mRNA synthesis accompanies loss of λ2 spikes from reovirus cores: an effect of λ2 on λ1 shell structure. Virology 296, 24–38 (2002)
Patton, J. T., Jones, M. T., Kalbach, A. N., He, Y.-W. & Xiaobo, J. Rotavirus RNA polymerase requires the core shell protein to synthesize the double-stranded RNA genome. J. Virol. 71, 9618–9626 (1997)
Mansell, E. A. & Patton, J. T. Rotavirus RNA replication: VP2, but not VP6, is necessary for viral replicase activity. J. Virol. 64, 4988–4996 (1990)
Gridley, C. L. & Patton, J. T. Regulation of rotavirus polymerase activity by inner capsid proteins. Curr. Opin. Virol. 9, 31–38 (2014)
McDonald, S. M. & Patton, J. T. Rotavirus VP2 core shell regions critical for viral polymerase activation. J. Virol. 85, 3095–3105 (2011)
Starnes, M. C. & Joklik, W. K. Reovirus protein λ3 is a poly (C)-dependent poly (G) polymerase. Virology 193, 356–366 (1993)
Reinisch, K. M., Nibert, M. L. & Harrison, S. C. Structure of the reovirus core at 3.6Å resolution. Nature 404, 960–967 (2000)
Grimes, J. M. et al. The atomic structure of the bluetongue virus core. Nature 395, 470–478 (1998)
Yu, X., Jin, L. & Zhou, Z. H. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature 453, 415–419 (2008)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Lyumkis, D., Brilot, A. F., Theobald, D. L. & Grigorieff, N. Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 183, 377–388 (2013)
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012)
Yu, X., Ge, P., Jiang, J., Atanasov, I. & Zhou, Z. H. Atomic model of CPV reveals the mechanism used by this single-shelled virus to economically carry out functions conserved in multishelled reoviruses. Structure 19, 652–661 (2011)
Zhang, X. et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc. Natl Acad. Sci. USA 105, 1867–1872 (2008)
Wolf, M., Garcea, R. L., Grigorieff, N. & Harrison, S. C. Subunit interactions in bovine papillomavirus. Proc. Natl Acad. Sci. USA 107, 6298–6303 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Huiskonen, J. T., Jaalinoja, H. T., Briggs, J. A., Fuller, S. D. & Butcher, S. J. Structure of a hexameric RNA packaging motor in a viral polymerase complex. J. Struct. Biol. 158, 156–164 (2007)
Briggs, J. A. et al. Classification and three-dimensional reconstruction of unevenly distributed or symmetry mismatched features of icosahedral particles. J. Struct. Biol. 150, 332–339 (2005)
Booy, F. et al. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 64, 1007–1015 (1991)
Zhang, Y., Kostyuchenko, V. A. & Rossmann, M. G. Structural analysis of viral nucleocapsids by subtraction of partial projections. J. Struct. Biol. 157, 356–364 (2007)
Tao, Y. et al. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell 95, 431–437 (1998)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Zhang, X. et al. A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 Å resolution. Elife 2, e01299 (2013)
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (AI094386 and GM071940 to Z.H.Z.), NSFC (31172263 to J.S.) and NSFGD (S2013010016750 to J.S.). We acknowledge the use of instruments at the Electron Imaging Center for Nanomachines supported by UCLA and by instrumentation grants from NIH (1S10RR23057, 1S10OD018111) and NSF (DBI-1338135). We thank P. Ge for carrying out a Relion reconstruction without an initial model as an independent verification step, S. Schein and L. Wang for proof-reading the paper, and P. Afonine for model refinement.
Author information
Authors and Affiliations
Contributions
Z.H.Z. supervised research; X.Z., X.Y. and Z.H.Z. designed and performed the experiments, analysed and interpreted data and wrote the paper; K.D. wrote programs, analysed data and prepared figures; W.C. built models; J.S. prepared reagents. All authors reviewed and finalized the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
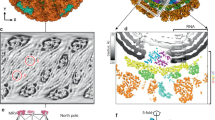
Extended Data Figure 1 Illustration of the asymmetric reconstruction procedure using particles with the capsid density subtracted.
See Methods for full explanation of panels a–m.
Extended Data Figure 2 Validation of asymmetric reconstruction from capsid-subtracted images using a Gaussian ball as the initial model.
Arrows linking a to f represent the progression of the procedure. The top panels (a, c, e) show the input model for each run and the bottom panels (b, d, f) show the output of each run.
Extended Data Figure 3 Sections of the q-CPV density map along the three-fold (that is, the earth axis) (a) and two-fold (b) axes of the pseudo-D3 symmetry.
Note the lack of three-fold and two-fold symmetry in the RNA density in contrast to the perfect symmetry of the capsid shell proteins. Pixel size = 4.04 Å; clipped map size = 166 × 166 × 120 pixels.
Extended Data Figure 4 dsRNA density maps in the quiescent state.
a, View of TEC + RNA densities with the same orientation of Fig. 1d. b,c, The same view as in a but rotated by +90° (b) or −90° (c) along x axis in panel a to view from either north (b) or south (c) poles. d–f, Three views from three two-fold axes on the equator, each is rotated by 120° along the y axis from each other. g, dsRNA density maps at the twelve vertices. TECs are arranged and numbered according to Fig. 1d. First row, TECs 1, 2, 3; second row, TECs 4, 5, 6; third row, TEC 7 and two unoccupied positions; fourth row, TECs 8, 9, 10. All TECs have a dsRNA segment bonded at the flange, each marked with a black arrow. Unlike the polar TECs, each tropical TEC (4–7) is surrounded by an extra density rod (open arrow).
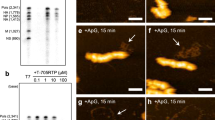
Extended Data Figure 5 CryoEM reconstructions of CPV in the quiescent and transcribing states.
a, b, CryoEM images of CPV particles in quiescent (a) and transcribing (b) states. These images were obtained by aligning and averaging frames in direct electron counting image stacks. Fibre-like nascent mRNAs are visible over background in b (marked by green arrows), while the background in a is clean. c, d, Fourier shell correlation coefficients (FSCs) as a function of spatial frequency between two half maps for reconstructions in the quiescent (c) and transcribing (d) states. The black and red lines represent FSCs for the asymmetrical reconstructions of capsid + genome and the locally averaged TEC densities, respectively. The effective resolutions of the local averaged maps are ~3.3 Å (c) and ~4.0 Å (d) resolution (FSC≥0.143) for maps in the quiescent and transcribing, respectively. e, f, CryoEM densities (grey surface representations) superimposed with atomic models (ribbons and sticks) for the quiescent (e) and transcribing (f) states. The α-helix (Pα12) and the four-stranded β-sheet (P4, P7–8 and P11) in e and f are both from the palm subdomain of the polymerase domain at 3.3 Å (e) and ~4.0 Å (f) resolutions. g, Statistics of CPV reconstructions and atomic model refinement.
Extended Data Figure 6 Sequence and secondary structure assignment of CPV RdRP in the quiescent state.
α-helices were marked by cylinders, β-strands by arrows, loops by thin lines, and the flexible tip domain by dashed lines. The colour scheme is the same as Fig. 3a.
Extended Data Figure 7 The RdRP-bound dsRNA in the quiescent and transcribing states.
a–c, Location of a TEC on the inner surface of the capsid shell in the quiescent and transcribing states. The inner surface of the CPV capsid (a) with 10 CSPs labelled (CSP A.1/B.1 to CSP A.5/B.5). b, c, Position of a TEC on the inner surface of capsid in the quiescent (b) and transcribing (c) states. VP4 and RdRP are coloured cyan and purple, respectively. An icosahedral five-fold axis is indicated with a small green pentagon. d, e, CryoEM densities of TEC and dsRNA (orange) in the quiescent (d) and transcribing (e) states. f, g, Models of TEC (surface representation) and dsRNA (ribbons) in the quiescent (f) and transcribing (g) states. Close-up views show the bound dsRNA (surface representation) on RdRP in the quiescent state (f) and its detachment in the transcribing state (g). VP4 is coloured cyan and the RdRP is coloured as in Fig. 3a. All surfaces displayed in this figure were rendered from models, except for the density maps of RdRP + dsRNA in d, e.
Extended Data Figure 8 Tracing amino acid residues 910–932 and 971–1000 of module B of the bracelet domain of RdRP in the quiescent and transcribing states.
a, b, CryoEM densities of RdRP in the quiescent (a) and transcribing (b) states. The locations of the residues 910–932 and 971–1000 are indicated with cyan boxes in a and b. Owing to their flexibility, these residues are not readily visible when displayed as in a and b but become visible when the maps are filtered to a lower resolution (for example, 4.5 Å resolution) as in c–f. The colour scheme of domains/subdomains is the same as in Fig. 3a. c, Trace of the residues 971–1000 (green) and 910–932 (purple) of module B of the bracelet domain of RdRP in the quiescent state. d, The same as c but in a different view. e, Trace of the residues 971–1000 (green) and 910–932 (purple) of module B of the bracelet domain of RdRP in the transcribing state. f, The same as e but in a different view to show the unambiguous trace of the two peptide fragments. g, h, Trace of the residues 910–923 (g) (purple) and 926–932 (h) (purple) of the bracelet domain of RdRP in the transcribing state, showing the unambiguous trace of the two peptide fragments. i, j, CryoEM densities (grey) and model (ribbon) of RdRP in the transcribing state, showing α-helices (i) and a β-hairpin (j). The colour scheme of domains/subdomains is the same as in Fig. 3a, k, l, Trace of the residues of the bracelet domain of RdRP in the transcribing state, showing a α-helix (k) and a β-sheet (l).
Extended Data Figure 9 Stereo and rotated views of Fig. 4a, b.
a, b, Stereo views of modules A (yellow cylinders and loops) and B (purple cylinders and loops) of the bracelet domain of RdRP in the quiescent (a) and transcribing (b) states. c, d, Same as in a, b, but rotated around the x axis by 90°. All surfaces displayed in this figure were rendered from models.
Extended Data Figure 10 Comparisons of RdRPs from CPV and MRV.
a, b, CryoEM in situ structure of the RdRP in t-CPV (a) and crystal structure of the MRV RdRP (b), both containing a RNA duplex in the active site. c–e, Superposition of domains of RdRPs from t-CPV (colour) and MRV (grey): N-terminal (c), polymerase (d) and bracelet (e) domains. f–h, Comparisons of modules A (yellow) and B (magenta) of the bracelet domain of RdRPs from q-CPV (f), t-CPV (g) and MRV (h). i–k, The same as in f–h, but with helices shown as cylinders, as in Fig. 4a, b.
Supplementary information
Asymmetrical reconstruction of q-CPV, showing capsid (grey) and dsRNA (radially coloured as in Fig.1a) densities.
Asymmetrical reconstruction of q-CPV, showing capsid (grey) and dsRNA (radially coloured as in Fig.1a) densities. (MP4 26395 kb)
Earth-like representation illustrating the locations of the ten TECs.
The earth’s north-south axis is the “pseudo 3-fold axis” (marked in Figure 1d). (MP4 21611 kb)
VP4 density map (grey) superimposed with the atomic model.
VP4 density map (grey) superimposed with the atomic model. (MP4 15939 kb)
VP4 density map (mesh) of the GTP-binding site superimposed with the atomic model of a GTP.
VP4 density map (mesh) of the GTP-binding site superimposed with the atomic model of a GTP. (MP4 32909 kb)
CryoEM density map (grey) of RdRP superimposed with its atomic model.
CryoEM density map (grey) of RdRP superimposed with its atomic model. (MP4 23876 kb)
High resolution features of an α-helix for validation of resolution.
A density map of the Pα;12 helix from the palm subdomain of the RdRP in the quiescent state. The density is shown as a semi-transparent surface (grey) and is superimposed with the atomic model (sticks). Note that the side chains of amino acid residues match the densities in the cryoEM map. (MP4 3975 kb)
High resolution features of a β-sheet for Validation of resolution.
A density map of the P-core, four-stranded β-sheet (P4, P7-8 & P11) from the palm subdomain of the RdRP in the quiescent state. The density is shown as a semi-transparent surface (grey) and is superimposed with the atomic model (sticks). (MP4 7845 kb)
Density map of the TEC in the quiescent state, with color code: VP4 (cyan) and RdRP (yellow→N-terminal; purple→ C-terminal bracelet; red→Palm; blue→finger; green→thumb subdomains).
The density map is shown as a semi-transparent surface (grey) and is superimposed with the atomic model (ribbons and sticks). (MP4 14387 kb)
Density map of the TEC in the transcribing state, with the same colour code as in the Supplementary Video 8.
Density map of the TEC in the transcribing state, with the same colour code as in the Supplementary Video 8. (MP4 13686 kb)
Comparison of the RdRP structures in the quiescent and transcribing states.
The dsRNA duplex model is also displayed. (MP4 25073 kb)
Rights and permissions
About this article
Cite this article
Zhang, X., Ding, K., Yu, X. et al. In situ structures of the segmented genome and RNA polymerase complex inside a dsRNA virus. Nature 527, 531–534 (2015). https://doi.org/10.1038/nature15767
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature15767
This article is cited by
-
Recent advances in rotavirus reverse genetics and its utilization in basic research and vaccine development
Archives of Virology (2021)
-
Translation of the long-term fundamental studies on viral DNA packaging motors into nanotechnology and nanomedicine
Science China Life Sciences (2020)
-
Reoviridae transcription is more than an open-and-shut case
Nature Structural & Molecular Biology (2019)
-
Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus
Nature (2019)
-
In situ structures of rotavirus polymerase in action and mechanism of mRNA transcription and release
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.