Abstract
Clustered regularly interspaced short palindromic repeat (CRISPR) loci and their associated (Cas) proteins provide adaptive immunity against viral infection in prokaryotes. Upon infection, short phage sequences known as spacers integrate between CRISPR repeats and are transcribed into small RNA molecules that guide the Cas9 nuclease to the viral targets (protospacers). Streptococcus pyogenes Cas9 cleavage of the viral genome requires the presence of a 5′-NGG-3′ protospacer adjacent motif (PAM) sequence immediately downstream of the viral target. It is not known whether and how viral sequences flanked by the correct PAM are chosen as new spacers. Here we show that Cas9 selects functional spacers by recognizing their PAM during spacer acquisition. The replacement of cas9 with alleles that lack the PAM recognition motif or recognize an NGGNG PAM eliminated or changed PAM specificity during spacer acquisition, respectively. Cas9 associates with other proteins of the acquisition machinery (Cas1, Cas2 and Csn2), presumably to provide PAM-specificity to this process. These results establish a new function for Cas9 in the genesis of prokaryotic immunological memory.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Barrangou, R. & Marraffini, L. A. CRISPR–Cas systems: prokaryotes upgrade to adaptive immunity. Mol. Cell 54, 234–244 (2014)
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007)
Deltcheva, E. et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607 (2011)
Makarova, K. S. et al. Evolution and classification of the CRISPR–Cas systems. Nature Rev. Microbiol. 9, 467–477 (2011)
Garneau, J. E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010)
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012)
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014)
Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA 109, E2579–E2586 (2012)
Szczelkun, M. D. et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl Acad. Sci. USA 111, 9798–9803 (2014)
Díez-Villaseñor, C., Guzman, N. M., Almendros, C., Garcia-Martinez, J. & Mojica, F. J. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR–Cas I-E variants of Escherichia coli. RNA Biol. 10, 792–802 (2013)
Yosef, I., Goren, M. G. & Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576 (2012)
Datsenko, K. A. et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 3, 945 (2012)
Nuñez, J. K. et al. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nature Struct. Mol. Biol. 21, 528–534 (2014)
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. A. RNA-guided editing of bacterial genomes using CRISPR–Cas systems. Nature Biotechnol. 31, 233–239 (2013)
Anders, C., Niewoehner, O., Duerst, A. & Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573 (2014)
Horinouchi, S. & Weisblum, B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150, 815–825 (1982)
Goldberg, G. W., Jiang, W., Bikard, D. & Marraffini, L. A. Conditional tolerance of temperate phages via transcription-dependent CRISPR–Cas targeting. Nature 514, 633–637 (2014)
Kreiswirth, B. N. et al. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712 (1983)
Bae, T., Baba, T., Hiramatsu, K. & Schneewind, O. Prophages of Staphylococcus aureus Newman and their contribution to virulence. Mol. Microbiol. 62, 1035–1047 (2006)
Sapranauskas, R. et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39, 9275–9282 (2011)
Li, M., Wang, R., Zhao, D. & Xiang, H. Adaptation of the Haloarcula hispanica CRISPR–Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 42, 2483–2492 (2014)
Richter, C. et al. Priming in the Type I-F CRISPR–Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 42, 8516–8526 (2014)
Horvath, P. et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190, 1401–1412 (2008)
Fonfara, I. et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR–Cas systems. Nucleic Acids Res. 42, 2577–2590 (2014)
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004)
Wiedenheft, B. et al. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure 17, 904–912 (2009)
Jinek, M. et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 343, 1247997 (2014)
Arslan, Z., Hermanns, V., Wurm, R., Wagner, R. & Pul, U. Detection and characterization of spacer integration intermediates in type I-E CRISPR–Cas system. Nucleic Acids Res. 42, 7884–7893 (2014)
Horinouchi, S. & Weisblum, B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J. Bacteriol. 150, 804–814 (1982)
Duplessis, M. & Moineau, S. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 41, 325–336 (2001)
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods 6, 343–345 (2009)
Bikard, D. et al. Exploiting CRISPR–Cas nucleases to produce sequence-specific antimicrobials. Nature Biotechnol. 32, 1146–1150 (2014)
Moore, S. D., Prevelige, P. E. & Jr A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein. J. Virol. 76, 10245–10255 (2002)
Arslan, Z. et al. Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2. Nucleic Acids Res. 41, 6347–6359 (2013)
Acknowledgements
We thank members of the laboratory for critical discussion of the experiments and their results, A. Zaytsev for help with the deep sequencing data analysis and A. Sherlock for help with plasmid construction. R.H. is the recipient of a Howard Hughes International Student Research Fellowship. P.S. is supported by a Helmsley Postdoctoral Fellowship for Basic and Translational Research on Disorders of the Digestive System at The Rockefeller University. J.W.M. is a Fellow of The Jane Coffin Childs Memorial Fund for Medical Research. D.B. is supported by a Harvey L. Karp Discovery Award and the Bettencourt Schuller Foundation. L.A.M. is supported by the Rita Allen Scholars Program, an Irma T. Hirschl Award, a Sinsheimer Foundation Award and a NIH Director’s New Innovator Award (1DP2AI104556-01).
Author information
Authors and Affiliations
Contributions
R.H., P.S., D.B. and L.A.M. conceived the study and designed experiments. R.H. and P.S. executed the experimental work with help from C.W. J.W.M. set up the experimental system to detect spacer acquisition in the absence of phage infection. G.W.G. isolated and characterized phage ϕNM4γ4 and constructed the pGG32 plasmid. D.B. analysed MiSeq data. L.A.M. wrote the paper with the help of the rest of the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
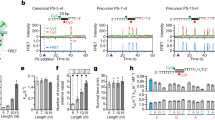
Extended Data Figure 1 The S. pyogenes type II CRISPR–Cas system displays a strong bias for the acquisition of spacers matching viral protospacers with NGG PAMs.
a, Analysis of bacteriophage-insensitive mutant colonies using PCR and agarose gel electrophoresis, representative of five technical replicates. Bacteria and phage were mixed in top agar and incubated overnight. DNA was isolated from individual colonies resistant to phage infection and used as template for a PCR reaction with primers (arrows) H182 and H183 (Extended Data Table 4), which amplify the 5′ end of the S. pyogenes CRISPR array. The size of the PCR band indicates the number of new spacers (shown at the top of the gel). Cells without additional spacers resist infection by a CRISPR-independent mechanisms, presumably envelope resistance. b, Analysis of acquired spacers during phage infection of a population of bacteria carrying the S. pyogenes type II CRISPR–Cas system. Liquid cultures of bacteria were infected with phage, surviving cells were collected at the end of the infection, DNA extracted and used as template for a PCR reaction as described above. Amplification products were separated by agarose gel electrophoresis and the DNA of the bands corresponding to products with additional spacers was extracted and sent for Mi-Seq next-generation sequencing. Reads corresponding to newly acquired spacers were plotted according to their position in the phage ϕNM4γ4 genome (x axis) and their abundance (y axis). Each dot represents a unique spacer sequence; blue and red dots indicate a corresponding protospacer with an NGG or non-NGG PAM. Top and bottom plots indicate protospacers in the top and bottom strands of the ϕNM4γ4 DNA. The map as well as the different functions of the phage genes are indicated in between the plots. The raw data used to make this graph is in the Source Data File . c, Weblogo showing the conservation of the 5′ flanking sequences of 10,000 protospacers randomly selected from the experiment shown in b. Absolute conservation of the NGG PAM was observed.
Extended Data Figure 2 cas1, cas2 and csn2 are not required for the execution of immunity.
a, Analysis of bacteriophage-resistant mutants that do not acquire a new spacer. Three colonies that survived phage infection in our in-plate adaptation assay (Extended Data Fig. 1) were subjected to phage adsorption assay. Briefly, surviving colonies as well as the wild-type S. aureus RN4220 control were grown in liquid and mixed with bacteriophage. After a brief incubation, cells were pelleted by centrifugation and the phages present in the supernatant (unable to bind and infect cells) were counted on a lawn of sensitive cells. The number of plaque-forming units (p.f.u.) of a control experiment in the absence of host cells were used to determine the 100% free-phage, or 0% adsorption value. No plaques were observed in the control experiment using wild-type cells and this value was used to set the 100% adsorption limit. The three CRISPR-independent, bacteriophage-resistant mutants displayed a marked defect in phage adsorption (about 50%), indicating that most likely they carry envelope resistance mutations. Error bars: mean ± s.d. (n = 3). b, cas1, cas2 and csn2 are not required for the execution of immunity using previously acquired spacers. Position within the phage ϕNM4γ4 genome of the type II CRISPR–Cas target used in this experiment. The protospacer sequence is in the bottom strand (shown in 3′–5′ direction) and flanked by a TGG PAM (in green). c, Comparison of immunity provided by a type II CRISPR–Cas system programmed to target the sequence shown in panel a in the presence (wild-type, wt) or absence (Δcas1,Δcas2, Δcsn2) of cas1, cas2 and csn2. Immunity is measured as the p.f.u. of a ϕNM4γ4 phage lysate spotted on top agar lawns of S. aureus RN4220 cells containing no CRISPR system (−), a wild-type S. pyogenes CRISPR–Cas type II system (wt, pRH233), or the same CRISPR–Cas systems with a deletion of cas1, cas2 and csn2 genes (Δcas1, Δcas2, Δcsn2, pRH079). Error bars: mean ± s.d. (n = 3).
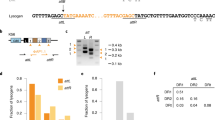
Extended Data Figure 3 Generation of an experimental system for the overexpression of cas1, cas2 and csn2 and the detection of spacer acquisition in the absence of phage infection.
a, Plasmids used in the spacer acquisition experiments presented in Figs 1c and 2c, d. pRH223 contains cas1, cas2 and csn2 from S. pyogenes under a tetracycline-inducible promoter. Cells containing this plasmid only acquired spacers when a second plasmid expressing cas9 was introduced, pRH240 or pRH241, containing the tracrRNA gene, the leader and first repeat from the S. pyogenes type II CRISPR–Cas system as well as cas9 from S. pyogenes (cas9Sp) or S. thermophilus (cas9St), respectively. The leader is a short, AT-rich sequence immediately upstream of the first repeat that contains the promoter for the transcription of the CRISPR array. b, Highly sensitive PCR assay to enrich for amplification products of adapted CRISPR loci. Arrows indicate primer annealing position and direction. The forward primer (JW8) anneals on the leader. For the reverse primer, a cocktail of JW3, JW4 and JW5 was used. The three reverse primers anneal on the repeat and differ only in their 3′-end nucleotide that never matches the last nucleotide of the leader (red arrowhead). Because this nucleotide is critical for the annealing of the primers, loci that acquire spacers ending in A, C or T are preferentially amplified over unadapted loci. c, To quantify the sensitivity of this technique, we mixed pGG32 (one repeat, unadapted) with pRH087 (repeat-spacer-repeat, adapted) in known ratios. The amplification of adapted plasmid was detected even when it represented 0.01% (10−4) of the total plasmid template, representative of three technical replicates. This highly sensitive PCR assay is not required to detect acquisition during phage infection, as in this case adapted cells survive and are enriched within the population, making their detection much easier.
Extended Data Figure 4 Purification of a Cas9–Cas1–Cas2–Csn2 complexes.
a, The cas9–cas1–cas2–csn2 operon of S. pyogenes SF370 was cloned into the pET16b vector (generating pKW07) to add an N-terminal histidyl tag to Cas9 and express all proteins in E. coli. Purification was performed using Ni-NTA affinity chromatography. SDS–PAGE followed by Coomassie staining of the purified proteins revealed a co-purifying protein that was identified as Cas1 by mass spectrometry, in a result representative of five technical replicates. Mass spectrometry identification of all the eluted proteins co-purifying with Cas9 is shown in Extended Data Table 2. b, The cas9–cas1–cas2–csn2 operon of S. pyogenes SF370 was cloned into the pET23a vector (generating pKW06) to add an C-terminal histidyl tag to Csn2 and express all proteins in E. coli. Purification was performed using Ni-NTA affinity chromatography followed by ion exchange chromatography. The elution fractions that constituted the peak containing the complex (Fig. 3a) were separated by SDS–PAGE and visualized by Coomassie staining, representative of three technical replicates.
Extended Data Figure 5 dCas9St can also support spacer acquisition.
A plasmid derived from pRH241 containing mutations in the active site of S. thermophilus Cas9 (D10A, H847A; dCas9St) was used to characterize spacer acquisition in the absence of phage infection. Upon overexpression of Cas1, Cas2 and Csn2 using anydrotetracycline (aTc), we were able to detect spacer acquisition. Sequencing of spacers and alignment of the protospacer flanking sequences demonstrated the selection of an NGGNG PAM. The image is representative of three technical replicates.
Extended Data Figure 6 A model for the selection of PAM-flanking spacers by Cas9.
After injection of the phage DNA, an adaptation complex formed by Cas9, Cas1, Cas2 and Csn2 uses the Cas9 PAM binding domain to specify functional protospacers, that is, that are followed by the correct PAM. It is not known how the protospacer sequence is extracted from the viral DNA to become a spacer. In the ‘cut and paste’ model, a nuclease, possibly Cas1, cuts the viral DNA to generate the spacer. In the ‘copy and paste’ model the protospacer sequence is copied first. Once loaded with the selected protospacer sequence, this complex promotes the integration of this sequence into the CRISPR array, thus becoming a new spacer. Previous studies demonstrated that Cas1 dimerizes and interacts with Cas2 (ref. 13); Csn2 has been determined to form a tetramer34.
Source data
Rights and permissions
About this article
Cite this article
Heler, R., Samai, P., Modell, J. et al. Cas9 specifies functional viral targets during CRISPR–Cas adaptation. Nature 519, 199–202 (2015). https://doi.org/10.1038/nature14245
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14245
This article is cited by
-
Co-delivery of Cas9 mRNA and guide RNAs for editing of LGMN gene represses breast cancer cell metastasis
Scientific Reports (2024)
-
Histones direct site-specific CRISPR spacer acquisition in model archaeon
Nature Microbiology (2023)
-
CRISPR-Cas provides limited phage immunity to a prevalent gut bacterium in gnotobiotic mice
The ISME Journal (2023)
-
Insertion sequence transposition inactivates CRISPR-Cas immunity
Nature Communications (2023)
-
Bacterial cGAS senses a viral RNA to initiate immunity
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.