Abstract
Deregulation of lysine methylation signalling has emerged as a common aetiological factor in cancer pathogenesis, with inhibitors of several histone lysine methyltransferases (KMTs) being developed as chemotherapeutics1. The largely cytoplasmic KMT SMYD3 (SET and MYND domain containing protein 3) is overexpressed in numerous human tumours2,3,4. However, the molecular mechanism by which SMYD3 regulates cancer pathways and its relationship to tumorigenesis in vivo are largely unknown. Here we show that methylation of MAP3K2 by SMYD3 increases MAP kinase signalling and promotes the formation of Ras-driven carcinomas. Using mouse models for pancreatic ductal adenocarcinoma and lung adenocarcinoma, we found that abrogating SMYD3 catalytic activity inhibits tumour development in response to oncogenic Ras. We used protein array technology to identify the MAP3K2 kinase as a target of SMYD3. In cancer cell lines, SMYD3-mediated methylation of MAP3K2 at lysine 260 potentiates activation of the Ras/Raf/MEK/ERK signalling module and SMYD3 depletion synergizes with a MEK inhibitor to block Ras-driven tumorigenesis. Finally, the PP2A phosphatase complex, a key negative regulator of the MAP kinase pathway, binds to MAP3K2 and this interaction is blocked by methylation. Together, our results elucidate a new role for lysine methylation in integrating cytoplasmic kinase-signalling cascades and establish a pivotal role for SMYD3 in the regulation of oncogenic Ras signalling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
11 June 2014
Affiliation 4 has been updated.
References
Helin, K. & Dhanak, D. Chromatin proteins and modifications as drug targets. Nature 502, 480–488 (2013)
Watanabe, T. et al. Differential gene expression signatures between colorectal cancers with and without KRAS mutations: crosstalk between the KRAS pathway and other signalling pathways. Eur. J. Cancer 47, 1946–1954 (2011)
Gaedcke, J. et al. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosom. Cancer 49, 1024–1034 (2010)
Hamamoto, R. et al. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nature Cell Biol. 6, 731–740 (2004)
Pylayeva-Gupta, Y., Grabocka, E. & Bar-Sagi, D. RAS oncogenes: weaving a tumorigenic web. Nature Rev. Cancer 11, 761–774 (2011)
Van Aller, G. S. et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics 7, 340–343 (2012)
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001)
Hingorani, S. R. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003)
Zhu, L., Shi, G., Schmidt, C. M., Hruban, R. H. & Konieczny, S. F. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am. J. Pathol. 171, 263–273 (2007)
Guerra, C. et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 11, 291–302 (2007)
Means, A. L. et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development 132, 3767–3776 (2005)
Morris, J. P. t., Cano, D. A., Sekine, S., Wang, S. C. & Hebrok, M. β-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J. Clin. Invest. 120, 508–520 (2010)
Bardeesy, N. et al. Both p16Ink4a and the p19Arf-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc. Natl Acad. Sci. USA 103, 5947–5952 (2006)
Feldser, D. M. et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 468, 572–575 (2010)
Junttila, M. R. et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature 468, 567–571 (2010)
Johnson, L. et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 (2001)
Levy, D. et al. A proteomic approach for the identification of novel lysine methyltransferase substrates. Epigenetics Chromatin 4, 19 (2011)
Maruyama, T. et al. CHIP-dependent termination of MEKK2 regulates temporal ERK activation required for proper hyperosmotic response. EMBO J. 29, 2501–2514 (2010)
Sun, W. et al. MEK kinase 2 and the adaptor protein Lad regulate extracellular signal-regulated kinase 5 activation by epidermal growth factor via Src. Mol. Cell. Biol. 23, 2298–2308 (2003)
Fanger, G. R., Johnson, N. L. & Johnson, G. L. MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J. 16, 4961–4972 (1997)
Choi, M. C. et al. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol. Cell 47, 122–132 (2012)
Matitau, A. E., Gabor, T. V., Gill, R. M. & Scheid, M. P. MEKK2 kinase association with 14-3-3 protein regulates activation of c-Jun N-terminal kinase. J. Biol. Chem. 288, 28293–28302 (2013)
Enomoto, A. et al. Negative regulation of MEKK1/2 signaling by serine-threonine kinase 38 (STK38). Oncogene 27, 1930–1938 (2008)
Brunet, A., Pages, G. & Pouyssegur, J. Constitutively active mutants of MAP kinase kinase (MEK1) induce growth factor-relaxation and oncogenicity when expressed in fibroblasts. Oncogene 9, 3379–3387 (1994)
Raman, M., Chen, W. & Cobb, M. H. Differential regulation and properties of MAPKs. Oncogene 26, 3100–3112 (2007)
Eichhorn, P. J., Creyghton, M. P. & Bernards, R. Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta 1795, 1–15 (2009)
Li, Y. M. & Casida, J. E. Cantharidin-binding protein: identification as protein phosphatase 2A. Proc. Natl Acad. Sci. USA 89, 11867–11870 (1992)
Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nature Genet. 29, 418–425 (2001)
Kawaguchi, Y. et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nature Genet. 32, 128–134 (2002)
Hruban, R. H. et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 66, 95–106 (2006)
DuPage, M., Dooley, A. L. & Jacks, T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature Protocols 4, 1064–1072 (2009)
Winslow, M. M. et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011)
Tiscornia, G., Singer, O. & Verma, I. M. Production and purification of lentiviral vectors. Nature Protocols 1, 241–245 (2006)
Heid, I. et al. Early requirement of Rac1 in a mouse model of pancreatic cancer. Gastroenterology 141, 719–730 (2011)
Mazur, P. K. et al. Notch2 is required for progression of pancreatic intraepithelial neoplasia and development of pancreatic ductal adenocarcinoma. Proc. Natl Acad. Sci. USA 107, 13438–13443 (2010)
Khatri, P. et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J. Exp. Med. 210, 2205–2221 (2013)
Storey, J. D. A direct approach to false discovery rates. J. R. Stat. Soc. Series B 64, 479–498 (2002)
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2003)
Brown, M. A., Sims, R. J., III, Gottlieb, P. D. & Tucker, P. W. Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol. Cancer 5, 26 (2006)
Green, E. M., Mas, G., Young, N. L., Garcia, B. A. & Gozani, O. Methylation of H4 lysines 5, 8 and 12 by yeast Set5 calibrates chromatin stress responses. Nature Struct. Mol. Biol. 19, 361–363 (2012)
Grey, C. et al. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol. 9, e1001176 (2011)
Kuo, A. J. et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell 44, 609–620 (2011)
Levy, D. et al. Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-κB signaling. Nature Immunol. 12, 29–36 (2011)
Moore, K. E. et al. A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol. Cell 50, 444–456 (2013)
Saddic, L. A. et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. 285, 37733–37740 (2010)
Shi, X. et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol. Cell 27, 636–646 (2007)
Strahl, B. D. et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22, 1298–1306 (2002)
Tachibana, M., Sugimoto, K., Fukushima, T. & Shinkai, Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276, 25309–25317 (2001)
Acknowledgements
We thank members of the Gozani and Sage laboratories for critical reading of the manuscript, A. Smits for help with mass spectrometry data visualization and P.J. Utz and the Floren Family Trust for providing ProtoArrays. This work was supported in part by grants from the NIH to O.G. and J.S. (RO1 CA172560) and an NIH Innovator grant (DP2 OD007447) from the Office of the Director for B.A.G.; M.V. was supported by a grant from NWO-VIDI, P.K.M. was supported by the Tobacco-Related Disease Research Program, a Dean’s Fellowship from Stanford University, and the Child Health Research Institute and Lucile Packard Foundation for Children’s Health at Stanford. N.R. was supported by a grant from the Fondation pour la Recherche Médicale. J.S. is the Harriet and Mary Zelencik Scientist in Children's Cancer and Blood Diseases.
Author information
Authors and Affiliations
Contributions
N.R. and P.K.M. contributed equally to this work and are listed alphabetically. They were responsible for the experimental design, execution, data analysis and manuscript preparation. P.K. and A.J.B. performed the bioinformatics meta-analysis. P.W.T.C.J. and M.V. performed the SILAC experiments. S.L. and B.A.G. performed the methylated peptide mass spectrometry experiments. A.W.W. generated recombinant H3 and H3K4R protein. O.B., G.S.V., M.H., D.D., P.J.T. and R.G.K. generated SMYD3 and MAP3K2me antibodies, the MAP3K2 peptides, and determined the catalytic efficiency of SMYD3. O.G. and J.S. were equally responsible for supervision of research, data interpretation and manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
O.G. is a co-founder of EpiCypher, Inc. O.B., G.S.V., M.H., D.D., P.J.T. and R.G.K. are employees of GSK.
Extended data figures and tables
Extended Data Figure 1 SMYD3 is a highly overexpressed KMT in Ras-associated cancers.
a, Analysis of seven publically available human pancreatic ductal adenocarcinoma (PDAC) gene expression studies from the NCBI GEO and EBI ArrayExpress for SMYD3 levels. The red line indicates expression of SMYD3 in pancreatic cancer biopsies (n = 203); the blue line marks normal pancreas samples (n = 91). The scale shows relative expression levels (log2). b, A bioinformatics meta-analysis identified 5 lysine methyltransferase overexpressed in human pancreatic ductal adenocarcinoma (PDAC). Meta-effect size and statistical tools are described in the Methods. FDR, false discovery rate. c, d, Summary of SMYD3 expression levels in seven (n = 294 independent samples) publicly-available expression data sets of PDAC and six data sets (n = 319 tumours and n = 147 normal independent samples) of non-small cell lung cancer (NSCLC), respectively. Detailed statistical description in the Methods section.
Extended Data Figure 2 Analysis of SMYD3 expression in human and mouse PDAC and lung adenocarcinoma (LAC).
a, Immunohistochemical analysis of SMYD3 expression in mouse and human WT pancreas, PanIN lesions, and PDAC. The expression pattern was further analysed using a Smyd3LacZ reporter knock-in strain. Smyd3LacZ mice were crossed to p48;KrasG12D (Kras) mice and studied at progressing stages of disease. Analysis of LacZ activity by X-gal staining as a surrogate for Smyd3 expression is shown (lower panel) (see Extended Data Fig. 3 for a cartoon of the knock-in allele). b, Immunoblot analysis with the indicated antibodies on tumour biopsy lysates from wild-type pancreas and from the pancreas of Kras mutant mice at 4.5 and 9 months of age when mice develop PanIN and PDAC, respectively (each time point represents two biological replicates). c, IHC analysis of SMYD3 expression in normal lung, atypical adenomatous hyperplasia (AAH), and lung adenocarcinoma (LAC). X-gal analysis of LacZ activity in Kras-driven tumours with the Smyd3LacZ reporter strain (lower panel). All images shown are representative. Arrowheads indicate nuclear localization of SMYD3. Scale bars, 50 μm. d, Smyd3 knockout allele diagram. In this allele, insertion of a LacZ cassette with a strong splice acceptor in intron 2 of the Smyd3 gene creates a mutant allele serving additionally as a reporter (Smyd3LacZ). Expression of the Cre recombinase in cells removes the LacZ cassette and further deletes Smyd3 exon 2, resulting in a null allele Smyd3KO. SA, splice acceptor; pA, polyadenylation signal.
Extended Data Figure 3 Smyd3 deletion inhibits pancreatic tumorigenesis.
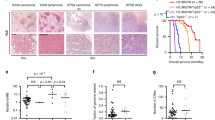
a, Analysis of pancreatic tumorigenesis at 6 months in Kras and Kras;Smyd3 mutant mice. Representative serial histology section (HE), IHC for pERK1/2 and the PanIN marker MUC5. b, Pancreatic cancer phenotypes in Kras;p53 and Kras;p53;Smyd3 mutant mice. Representative IHC for pERK1/2. Arrowheads indicate areas with intact acinar cells. c, Quantification of intact normal acinar area (amylase-positive area) in Kras;p53 and Kras;p53;Smyd3 mutant mice. Data are represented as mean ± s.e.m. ***P value < 0.001 (two-tailed unpaired Student’s t-test). d, Representative HE and pERK1/2 IHC images of lung sections from Kras and Kras;Smyd3 mutant mice 12, 16 and 20 weeks after Ad-Cre infection. pERK1/2 is a marker of Ras activity and advanced tumours. e, f, IHC analysis of SMYD3 expression in the PDAC (e) and LAC (h) mouse models. Arrowheads indicate cytoplasmic localization of SMYD3. Scale bars, 50 μm. g, h, Immunoblot analysis with the indicated antibodies probing pancreatic adenocarcinoma (g) or lung adenocarcinoma tumour lysates (h) dissected from Kras and Kras;Smyd3 mutant mice. Active Ras corresponds to Ras protein in the GTP-bound state pulled down with the RAF Ras-binding domain (RBD) (see Methods). *Tubulin loading control as in Fig. 1j and 2f, respectively.
Extended Data Figure 4 SMYD3 functions to maintain the tumorigenic characteristics of human and murine cancer cells.
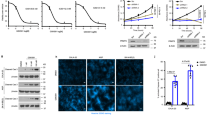
a–c, Cell proliferation rates (top panels) and colony formation in soft agar assays (bottom panels) of murine LAC cell line LKR10 (a), human LAC cell line A549 (b), or human PDAC cell line CFPac1 (c) with or without SMYD3 depletion by stable shRNA (respective immunoblot in middle panels). d–f, SMYD3 depletion in CFPac1 attenuates tumour growth in mouse xenografts. d, Macroscopic picture of xenografts from control and SMYD3 knock-down tumours at the end of the experiment. Scale bar, 1 cm. e, Volume analysis shows that shSMYD3 significantly inhibits the expansion of pancreatic tumours (n = 6 for each group). f, HE of the tumours and IHC confirmation of SMYD3 expression and knock-down. All scale bars, 50 μm. *P value < 0.05; **P value < 0.01; ***P value < 0.001 (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m.
Extended Data Figure 5 Lentiviral reconstitution of SMYD3 in pancreatic acinar-to-ductal-metaplasia (ADM) assays and in lung cancer cells in vivo.
a, IHC analysis of SMYD3 reconstitution in the lung (from Fig. 3a). b, Immunofluorescent detection of SMYD3 expression in wild-type and transduced acinar clusters (left panel). Acini (asterisk) transduced with lenti-Cre carrying wild-type SMYD3 but not catalytically inactive SMYD3(F183A) undergo ADM and form ducts (arrowhead) ex vivo. c, Quantification of acinar and ductal clusters after lentiviral infection (each treatment represents four independent biological replicates). Data are represented as mean ± s.e.m. *P value < 0.05 (two-tailed unpaired Student’s t-test).
Extended Data Figure 6 SMYD3 specifically methylates MAP3K2 at lysine 260 in vitro.
a, SMYD3 methylates MAP3K2 on protein arrays. Representative image (n = 3 independent experiments) showing a SMYD3 methylation assay on a ProtoArray. The close-up shows the two independent MAP3K2 spots on the array being methylated. b, SMYD3 is detected in the cytoplasm and not the nucleus in LKR10 cells. Immunoblot analysis with the indicated antibodies of LKR10 cell lysates biochemically separated into cytoplasmic, nuclear and chromatin fractions (see Methods). c, SMYD3 catalytic efficiency is two orders of magnitude greater on MAP3K2 than on H4. kcat, KM, and kcat/KM values of SMYD3 activity on recombinant H4 and MAP3K2 as substrates are shown. d, Schematic of the H3K4* mutant form used in e. Note that the only lysine available to be methylated in H3 is present at K4. e, In vitro methylation assay on full-length recombinant MAP3K2, H3 or H3K4* with recombinant SMYD3 and PRDM9. Top panels, short and long exposure autoradiograms of the methylation assay. No signal was detected for SMYD3 on H3 and H3K4* after long exposures. The asterisk and line indicate breakdown products of MAP3K2 that contain K260 and can be detected in this methylation assay upon long exposure. Bottom panel, Coomassie stain of proteins in the reaction. f, Positive control of activity for enzymes used in Fig. 3f, g on their known respective substrates (MAP3K2, histone H3, nucleosome or RelA as indicated). g, In vitro methylation assays using MAP3K2-K260meo, me1, me2 or me3 peptides as SMYD3 substrates. Dot blot is shown as control of peptide’s comparable concentration used for the methylation assay. h, Mass spectrometry analysis of SMYD3 methylation activity on unmodified MAP3K2-K260 peptide. i, Specificity of the indicated MAP3K2-K260me antibodies in dot blot assays using MAP3K2-K260meo, me1, me2 or me3 peptides. j, MAP3K2 is methylated in cells upon SMYD3 overexpression. Immunoblot analysis with the indicated antibodies from 293T cells lysates after Flag immunoprecipitation in cells overexpressing Flag–MAP3K2 and/or HA–SMYD3.
Extended Data Figure 7 SMYD3 and MAP3K2 knockdown both impair MAP kinase signalling.
a–d, Immunoblot analysis with the indicated antibodies of LKR10 (a, b), A549 (c), and CFPac1 (d) lysates. Asterisk indicates a slower migrating ERK5 species that is phosphorylated. Stimulation, 10% serum-complemented media for 15 min (b) or EGF for 15 min at 25 ng μl−1 (a, c, d). Immunoblots are representative of 3 independent biological replicates. e, f, SMYD3 methylation of MAP3K2 does not alter the intrinsic kinase activity of MAP3K2. e, Immunoblot analysis with the indicated antibodies from lysates of 293T cells transfected with control vector, wild-type SMYD3, catalytically dead SMYD3(F183A), wild-type MAP3K2, MAP3K2(K260A), or kinase dead MAP3K2(K385M). f, Methylation of MAP3K2 does not alter its in vitro kinase activity. In vitro kinase assays were performed with the indicated recombinant versions of MAP3K2 (wild-type, SMYD3-resistant K260A mutant, or kinase dead K385 mutant) pre-methylated with wild-type SMYD3 or as a control, inactive SMYD3, using MEK1 as a substrate. MEK1 phosphorylation was detected by immunoblot analysis with the indicated antibody.
Extended Data Figure 8 SMYD3 knockout augments the effects of the MEK1/2 inhibitor trametinib (GSK1120212) in vivo.
a, Schematic of the caerulein pancreatitis-induced tumorigenesis protocol. Mice were treated with a normal dose of trametinib (1 mg per kg intraperitoneally daily) or a low dose (0.1 mg per kg intraperitoneally daily) or vehicle control. b, Immunoblot analysis with indicated antibodies of two independent pancreas biopsies per treatment group. c, Quantification of MUC5-positive lesions in caerulein-treated pancreata from Kras and Kras;Smyd3 mutant mice treated with trametinib or vehicle control (n = 5, each treatment). *P value < 0.05; ***P value < 0.001 (two-tailed unpaired Student’s t-test). Data are represented as mean ± s.e.m. d, Representative macroscopic pictures of pancreata from each treatment group. Scale bar, 1 cm. e, Representative serial HE staining and IHC for pERK1/2, a marker of Ras activity, and MUC5, a marker of PanIN lesions. All scale bars, 50 μm.
Extended Data Figure 9 SMYD3 depletion augments the effects of the MEK1/2 inhibitor trametinib (GSK1120212) in Ras-driven cancer cells.
a–c, Relative cell proliferation rates (bottom panel) of murine LAC cell line LKR10 (a), human LAC cell line A549 (b), or human PDAC cell line CFPac1 (c) with or without SMYD3 depletion by stable shRNA (SMYD3 proteins levels are shown in top panel) in response to the indicated doses of trametinib. Experiments shown represent an average of 3 independent experiments performed in triplicates for each cancer line. Values represent the number of cells relative to control shRNA cells without treatment at 48 h. b, Constitutively active MEK1 (MEK1-DD) increases EGF-mediated ERK1/2 activation in SMYD3 depleted-cells. Immunoblot analysis with the indicated antibodies using lysates from A549 cells stably expressing shControl or shSMYD3 and transfected with HA–MEK1-DD. Stimulation: EGF treatment for 15 min at 25 ng μl−1.
Extended Data Figure 10 Treatment with the PP2A inhibitor cantharidin phenocopies SMYD3 function in vivo.
a, Schematic of the caerulein pancreatitis-induced tumorigenesis protocol. Mice were treated with the PP2A inhibitor cantharidin (iPP2A, 0.15 mg kg−1 intraperitoneally twice a day) or vehicle control. b, Immunoblot analysis with indicated antibodies on two independent pancreas biopsies per treatment group. c, Macroscopic pictures of WT and Kras;Smyd3 mutant pancreata. Note that treatment with the PP2A inhibitor leads to the development of enlarged, ‘hard’ pancreata characteristic of tumorigenic development even in Kras;Smyd3 mutant mice. Scale bar, 1 cm. d, Representative serial haematoxylin and eosin (HE) staining and IHC for pERK1/2, a marker of Ras activity, and MUC5, a marker of PanIN lesions. All scale bars, 50 μm. e, Summary model for SMYD3 regulation of MAP kinase signalling after MAP3K2 methylation. Oncogenic Ras activates several kinase cascades that play important roles in pancreas and lung cancer development, including four major MAPK pathways (ERK1/2, ERK5, JNK, and p38) as well as AKT signalling. SMYD3 is frequently overexpressed in pancreatic and lung cancers, two cancer types that are commonly driven by oncogenic Ras signalling. Overexpression of SMYD3 and the resulting methylation of MAP3K2 at K260 potentiate activation of kinases like ERK1/2 and ERK5 in response to stimuli like oncogenic Ras. We postulate a mechanism in which the PP2A complex is unable to bind methylated MAP3K2, which decreases the ability of this enzyme to terminate activating phosphorylation events on MAP3K2 and/or MAP3K2 downstream targets. Under conditions with excessive SMYD3 protein, the physiological relationship between PP2A and MAP3K2 is disrupted and results in an increased pathological MAP3K2 signalling, which cooperates with Ras to promote tumorigenesis.
Rights and permissions
About this article
Cite this article
Mazur, P., Reynoird, N., Khatri, P. et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 510, 283–287 (2014). https://doi.org/10.1038/nature13320
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13320
This article is cited by
-
SMYD3 promotes endometrial cancer through epigenetic regulation of LIG4/XRCC4/XLF complex in non-homologous end joining repair
Oncogenesis (2024)
-
Histone lysine methyltransferase SMYD3 promotes oral squamous cell carcinoma tumorigenesis via H3K4me3-mediated HMGA2 transcription
Clinical Epigenetics (2023)
-
A de novo PAK1 likely pathogenic variant and a de novo terminal 1q microdeletion in a Chinese girl with global developmental delay, severe intellectual disability, and seizures
BMC Medical Genomics (2023)
-
Methylation across the central dogma in health and diseases: new therapeutic strategies
Signal Transduction and Targeted Therapy (2023)
-
Inhibition of histone methyltransferase Smyd3 rescues NMDAR and cognitive deficits in a tauopathy mouse model
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.