Abstract
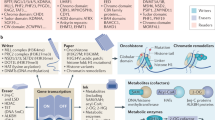
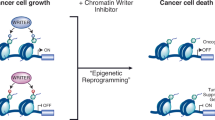
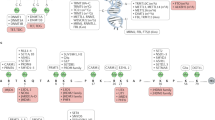
A plethora of groundbreaking studies have demonstrated the importance of chromatin-associated proteins and post-translational modifications of histones, proteins and DNA (so-called epigenetic modifications) for transcriptional control and normal development. Disruption of epigenetic control is a frequent event in disease, and the first epigenetic-based therapies for cancer treatment have been approved. A generation of new classes of potent and specific inhibitors for several chromatin-associated proteins have shown promise in preclinical trials. Although the biology of epigenetic regulation is complex, new inhibitors such as these will hopefully be of clinical use in the coming years.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Berger, S. L. The complex language of chromatin regulation during transcription. Nature 447, 407–412 (2007).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Baylin, S. B. & Jones, P. A. A decade of exploring the cancer epigenome — biological and translational implications. Nature Rev. Cancer 11, 726–734 (2011). This review provides an excellent overview of epigenetics with an emphasis on the linkage between genomic and epigenomic phenomena in cancer together with opportunities for biomarker-driven development of therapeutics.
Issa, J. P. & Kantarjian, H. M. Targeting DNA methylation. Clin. Cancer Res. 15, 3938–3946 (2009).
Yang, X., Lay, F., Han, H. & Jones, P. A. Targeting DNA methylation for epigenetic therapy. Trends Pharmacol. Sci. 31, 536–546 (2010).
Khan, O. & La Thangue, N. B. HDAC inhibitors in cancer biology: emerging mechanisms and clinical applications. Immunol. Cell Biol. 90, 85–94 (2012).
Wagner, J. M., Hackanson, B., Lubbert, M. & Jung, M. Histone deacetylase (HDAC) inhibitors in recent clinical trials for cancer therapy. Clin. Epigenetics 1, 117–136 (2010).
You, J. S. & Jones, P. A. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22, 9–20 (2012).
Huang, J. et al. p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 (2007).
Wu, S. C. & Zhang, Y. Role of protein methylation and demethylation in nuclear hormone signaling. Mol. Endocrinol. 23, 1323–1334 (2009).
Zhang, K. & Dent, S. Y. Histone modifying enzymes and cancer: going beyond histones. J. Cell. Biochem. 96, 1137–1148 (2005).
Robinson, G. et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48 (2012).
Ryan, R. J. & Bernstein, B. E. Genetic events that shape the cancer epigenome. Science 336, 1513–1514 (2012).
Shih, A. H., Abdel-Wahab, O., Patel, J. P. & Levine, R. L. The role of mutations in epigenetic regulators in myeloid malignancies. Nature Rev. Cancer 12, 599–612 (2012).
McCabe, M. T. et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc. Natl Acad. Sci. USA 109, 2989–2994 (2012).
Sneeringer, C. J. et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl Acad. Sci. USA 107, 20980–20985 (2010).
Kuo, A. J. et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell 44, 609–620 (2011).
Meyer, C. et al. New insights to the MLL recombinome of acute leukemias. Leukemia 23, 1490–1499 (2009).
Okada, Y. et al. hDOT1L links histone methylation to leukemogenesis. Cell 121, 167–178 (2005).
Bernt, K. M. et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20, 66–78 (2011).
Daigle, S. R. et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011). References 20 and 21 present compelling evidence for the involvement of DOT1L and H3K79 methylation in MLL-rearranged leukaemia and provide rationale for therapeutic targeting of the enzyme.
Anglin, J. L. et al. Synthesis and structure-activity relationship investigation of adenosine-containing inhibitors of histone methyltransferase DOT1L. J. Med. Chem. 55, 8066–8074 (2012).
Yu, W. et al. Bromo-deaza-SAH: a potent and selective DOT1L inhibitor. Bioorg. Med. Chem. 21, 1787–1794 (2013).
Daigle, S. R. et al. Potent inhibition of DOT1L as treatment for MLL-fusion leukemia. Blood http://dx.doi.org/10.1182/blood-2013-04-497644 (2013).
Morey, L. & Helin, K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 35, 323–332 (2010).
Varambally, S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002).
Dalgliesh, G. L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
van Haaften, G. et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nature Genet. 41, 521–523 (2009).
Kleer, C. G. et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl Acad. Sci. USA 100, 11606–11611 (2003).
Wagener, N. et al. Enhancer of zeste homolog 2 (EZH2) expression is an independent prognostic factor in renal cell carcinoma. BMC Cancer 10, 524 (2010).
Takawa, M. et al. Validation of the histone methyltransferase EZH2 as a therapeutic target for various types of human cancer and as a prognostic marker. Cancer Sci. 102, 1298–1305 (2011).
Bracken, A. P. et al. EZH2 is downstream of the pRB–E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22, 5323–5335 (2003).
Morin, R. D. et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature Genet. 42, 181–185 (2010).
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nature Genet. 43, 830–837 (2011).
Ryan, R. J. et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS ONE 6, e28585 (2011).
Nikoloski, G. et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nature Genet. 42, 665–667 (2010).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012). This is the first report of a potent and selective inhibitor of EZH2 with in vitro and in vivo activity compared with EZH2 mutant lymphomas.
Knutson, S. K. et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nature Chem. Biol. 8, 890–896 (2012).
Qi, W. et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl Acad. Sci. USA 109, 21360–21365 (2012).
Valente, S. et al. Identification of PR-SET7 and EZH2 selective inhibitors inducing cell death in human leukemia U937 cells. Biochimie 94, 2308–2313 (2012).
Epizyme Inc. Inhibitors of human EZH2, and methods to use thereof. WO/2012/034132 (2012).
Tan, J. et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 21, 1050–1063 (2007).
Miranda, T. B. et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 8, 1579–1588 (2009).
Knutson, S. K. et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl Acad. Sci. USA 110, 7922–7927 (2013). This article reports activity of EZH2 inhibitors in solid tumours suggesting potential for clinical benefit beyond haematological malignancies.
Kooistra, S. M. & Helin, K. Molecular mechanisms and potential functions of histone demethylases. Nature Rev. Mol. Cell Biol. 13, 297–311 (2012).
Mosammaparast, N. & Shi, Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79, 155–179 (2010).
Yang, Z. Q. et al. Identification of a novel gene, GASC1, within an amplicon at 9p23–24 frequently detected in esophageal cancer cell lines. Cancer Res. 60, 4735–4739 (2000).
Liu, G. et al. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene 28, 4491–4500 (2009).
Ehrbrecht, A. et al. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J. Pathol. 208, 554–563 (2006).
Lu, P. J. et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 274, 15633–15645 (1999).
Hayami, S. et al. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol. Cancer 9, 59 (2010).
He, J., Nguyen, A. T. & Zhang, Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood 117, 3869–3880 (2011).
Jensen, L. R. et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236 (2005).
Laumonnier, F. et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 42, 780–786 (2005).
Kruidenier, L. et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488, 404–408 (2012). This article describes the discovery, structural biology and activity of potent and selective Jumonji demethylase inhibitors.
Lynch, J. T., Harris, W. J. & Somervaille, T. C. LSD1 inhibition: a therapeutic strategy in cancer? Expert Opin. Ther. Targets 16, 1239–1249 (2012).
Hayami, S. et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int. J. Cancer 128, 574–586 (2011).
Kauffman, E. C. et al. Role of androgen receptor and associated lysine-demethylase coregulators, LSD1 and JMJD2A, in localized and advanced human bladder cancer. Mol. Carcinog. 50, 931–944 (2011).
Kahl, P. et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 66, 11341–11347 (2006).
Schulte, J. H. et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 69, 2065–2071 (2009).
Harris, W. J. et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell 21, 473–487 (2012).
Schenk, T. et al. Inhibition of the LSD1 (KDM1A) demethylase reactivates the all-trans-retinoic acid differentiation pathway in acute myeloid leukemia. Nature Med. 18, 605–611 (2012). Reference 61 and 62 highlight the role of LSD1 in AML and the potential for inhibitors to synergize with all- trans -retinoic acid therapy.
Oryzon Genomics. Phenylcyclopropylamine derivatives and their medical use. WO/2010/084160 (2010).
Ram, O. et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell 147, 1628–1639 (2011).
Whyte, W. A. et al. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482, 221–225 (2012).
Maes, T. et al. Preclinical characterization of a potent and selective inhibitor of the histone demethylase KDM1A for MLL leukemia. J. Clin. Oncol. 31, suppl; abstr. e13543 (2013).
Lee, M. G., Wynder, C., Schmidt, D. M., McCafferty, D. G. & Shiekhattar, R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 13, 563–567 (2006).
Zuccato, C. et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nature Genet. 35, 76–83 (2003).
Liang, Y., Vogel, J. L., Narayanan, A., Peng, H. & Kristie, T. M. Inhibition of the histone demethylase LSD1 blocks α-herpesvirus lytic replication and reactivation from latency. Nature Med. 15, 1312–1317 (2009).
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012).
Gyuris, A. et al. The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim. Biophys. Acta 1789, 413–421 (2009).
Nicodeme, E. et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 (2010).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Mitsubishi–Tanabe Pharma Corporation. Antitumor agent. WO/2009/084693 (2009).
French, C. A. Demystified molecular pathology of NUT midline carcinomas. J. Clin. Pathol. 63, 492–496 (2010).
Dawson, M. A. et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478, 529–533 (2011).
Delmore, J. E. et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). Together with references 72 and 73, references 76 and 77 are outstanding demonstrations of the feasibility of inhibiting bromodomain proteins in inflammation and tumorigenesis.
Puissant, A. et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 3, 308–323 (2013).
Béguelin, W. et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013).
Ernst, T. et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nature Genet. 42, 722–726 (2010).
Ntziachristos, P. et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nature Med. 18, 298–301 (2012).
Simon, C. et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 26, 651–656 (2012).
Zhang, J. et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481, 157–163 (2012).
Lewis, P. W. et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340, 857–861 (2013).
Stronach, E. A. et al. HDAC4-regulated STAT1 activation mediates platinum resistance in ovarian cancer. Cancer Res. 71, 4412–4422 (2011).
Yu, W. et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nature Commun. 3, 1288 (2012).
Amatangelo, M. D. et al. Three-dimensional culture sensitizes epithelial ovarian cancer cells to EZH2 methyltransferase inhibition. Cell Cycle 12, 2113–2119 (2013).
Structural Genomics Consortium. Chemical Probes. http://www.thesgc.org/scientists/chemical_probes (SGC, 2013).
Konze, K. D. et al. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem. Biol. 8, 1324–1334 (2013).
Kubicek, S. et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25, 473–481 (2007).
Liu, F. et al. Protein lysine methyltransferase G9a inhibitors: design, synthesis, and structure activity relationships of 2,4-diamino-7-aminoalkoxy-quinazolines. J. Med. Chem. 53, 5844–5857 (2010).
Vedadi, M. et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nature Chem. Biol. 7, 566–574 (2011).
Yuan, Y. et al. A small-molecule probe of the histone methyltransferase G9a induces cellular senescence in pancreatic adenocarcinoma. ACS Chem. Biol. 7, 1152–1157 (2012).
Liu, F. et al. Exploiting an allosteric binding site of PRMT3 yields potent and selective inhibitors. J. Med. Chem. 56, 2110–2124 (2013).
Wan, H. et al. Benzo[d]imidazole inhibitors of coactivator associated arginine methyltransferase 1 (CARM1) — hit to lead studies. Bioorg. Med. Chem. Lett. 19, 5063–5066 (2009).
Sack, J. S. et al. Structural basis for CARM1 inhibition by indole and pyrazole inhibitors. Biochem. J. 436, 331–339 (2011).
Allan, M. et al. N-Benzyl-1-heteroaryl-3-(trifluoromethyl)-1H-pyrazole-5-carboxamides as inhibitors of co-activator associated arginine methyltransferase 1 (CARM1). Bioorg. Med. Chem. Lett. 19, 1218–1223 (2009).
Seal, J. et al. Identification of a novel series of BET family bromodomain inhibitors: binding mode and profile of I-BET151 (GSK1210151A). Bioorg. Med. Chem. Lett. 22, 2968–2972 (2012).
Picaud, S. et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET bromodomains. Cancer Res. 73, 3336–3346 (2013).
Herold, J. M. et al. Small-molecule ligands of methyl-lysine binding proteins. J. Med. Chem. 54, 2504–2511 (2011).
James, L. I. et al. Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain. Nature Chem. Biol. 9, 184–191 (2013).
Metzger, E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 (2005).
Wissmann, M. et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nature Cell Biol. 9, 347–353 (2007).
Acknowledgements
Work in the Helin laboratory is supported by the Danish National Research Foundation, the Danish Cancer Society, the Novo Nordisk Foundation, the Lundbeck Foundation, the European Union, The European Research Council and the Excellence Programme of the University of Copenhagen.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
K.H. is a co-founder of EpiTherapeutic, and is a consultant for and has shares and warrants in the company. D.D. is a GSK shareholder and an employee of Jansen Pharmaceuticals.
Additional information
Reprints and permission information is available at www.nature.com/reprints.
Rights and permissions
About this article
Cite this article
Helin, K., Dhanak, D. Chromatin proteins and modifications as drug targets. Nature 502, 480–488 (2013). https://doi.org/10.1038/nature12751
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12751
This article is cited by
-
In-silico guided chemical exploration of KDM4A fragments hits
Clinical Epigenetics (2023)
-
Primary bilateral macronodular adrenal hyperplasia: definitely a genetic disease
Nature Reviews Endocrinology (2022)
-
SIX3 function in cancer: progression and comprehensive analysis
Cancer Gene Therapy (2022)
-
Oncogenic GBX2 promotes the malignant behaviors of bladder cancer cells by binding to the ITGA5 promoter and activating its transcription
Functional & Integrative Genomics (2022)
-
Diffuse intrinsic pontine glioma: current insights and future directions
Chinese Neurosurgical Journal (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.