Abstract
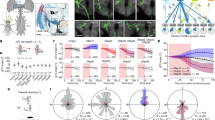
Animal behaviour arises from computations in neuronal circuits, but our understanding of these computations has been frustrated by the lack of detailed synaptic connection maps, or connectomes. For example, despite intensive investigations over half a century, the neuronal implementation of local motion detection in the insect visual system remains elusive. Here we develop a semi-automated pipeline using electron microscopy to reconstruct a connectome, containing 379 neurons and 8,637 chemical synaptic contacts, within the Drosophila optic medulla. By matching reconstructed neurons to examples from light microscopy, we assigned neurons to cell types and assembled a connectome of the repeating module of the medulla. Within this module, we identified cell types constituting a motion detection circuit, and showed that the connections onto individual motion-sensitive neurons in this circuit were consistent with their direction selectivity. Our results identify cellular targets for future functional investigations, and demonstrate that connectomes can provide key insights into neuronal computations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Heisenberg, M. & Wolf, R. Vision in Drosophila. Genetics of Microbehaviour (Springer Verlag, 1984)
Laughlin, S. B. Matching coding, circuits, cells, and molecules to signals: General principles of retinal design in the fly’s eye. Prog. Retin. Eye Res. 13, 165–196 (1994)
Strausfeld, N. J. & Nässel, D. R. in Handbook of Sensory Physiology (eds Autrum, H. et al.) 1–132 (Springer-Verlag, 1981)
Hassenstein, B. & Reichardt, W. Systemtheoretische Analyse der Zeit-, Reihenfolgen- und Vorzeichenauswertung bei der Bewegungsperzeption des Rüsselkäfers Chlorophanus. Z. Naturforsch. B 11, 513–524 (1956)
Reichardt, W. in Sensory Communication (ed. Rosenblith, W. A. ) 303–317 (MIT Press, 1961)
Barlow, H. B. & Levick, W. R. The mechanism of directionally selective units in rabbit's retina. J. Physiol. (Lond.) 178, 477–504 (1965)
Borst, A., Haag, J. & Reiff, D. F. Fly motion vision. Annu. Rev. Neurosci. 33, 49–70 (2010)
Rivera-Alba, M. et al. Wiring economy and volume exclusion determine neuronal placement in the Drosophila brain. Curr. Biol. 21, 2000–2005 (2011)
Meinertzhagen, I. A. & Sorra, K. E. Synaptic organization in the fly’s optic lamina: few cells, many synapses and divergent microcircuits. Prog. Brain Res. 131, 53–69 (2001)
Takemura, S. Y., Lu, Z. & Meinertzhagen, I. A. Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. J. Comp. Neurol. 509, 493–513 (2008)
Buchner, E., Buchner, S. & Bülthoff, I. Deoxyglucose mapping of nervous activity induced in Drosophila brain by visual movement. J. Comp. Physiol. A 155, 471–483 (1984)
Krapp, H. G. & Hengstenberg, R. Estimation of self-motion by optic flow processing in single visual interneurons. Nature 384, 463–466 (1996)
Joesch, M., Plett, J., Borst, A. & Reiff, D. F. Response properties of motion-sensitive visual interneurons in the lobula plate of Drosophila melanogaster. Curr. Biol. 18, 368–374 (2008)
White, J. G., Southgate, E., Thomson, J. N. & Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Phil. Trans. R. Soc. Lond. B 314, 1–340 (1986)
Kirschfeld, K. in Information Processing in the Visual System of Arthropods (ed. Wehner, R. ) 61–74 (Springer Verlag, 1972)
Riehle, A. & Franceschini, N. Motion detection in flies: parametric control over ON–OFF pathways. Exp. Brain Res. 54, 390–394 (1984)
Schuling, F. H., Mastebroek, H. A. K., Bult, R. & Lenting, B. P. M. Properties of elementary movement detectors in the fly Calliphora erythrocephala. J. Comp. Physiol. A 165, 179–192 (1989)
Helmstaedter, M., Briggman, K. L. & Denk, W. 3D structural imaging of the brain with photons and electrons. Curr. Opin. Neurobiol. 18, 633–641 (2008)
Chklovskii, D. B., Vitaladevuni, S. & Scheffer, L. K. Semi-automated reconstruction of neural circuits using electron microscopy. Curr. Opin. Neurobiol. 20, 667–675 (2010)
Fischbach, K.-F. & Dittrich, A. P. M. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 258, 441–475 (1989)
Song, S., Sjöström, P. J., Reigl, M., Nelson, S. & Chklovskii, D. B. Highly nonrandom features of synaptic connectivity in local cortical circuits. PLoS Biol. 3, e68 (2005)
Varshney, L. R., Chen, B. L., Paniagua, E., Hall, D. H. & Chklovskii, D. B. Structural properties of the Caenorhabditis elegans neuronal network. PLOS Comput. Biol. 7, e1001066 (2011)
Campos-Ortega, J. A. & Strausfeld, N. J. in Information Processing in the Visual Systems of Arthropods (ed. Wehner, R. ) 31–36 (Springer Verlag, 1972)
Franceschini, N., Kirschfeld, K. & Minke, B. Fluorescence of photoreceptor cells observed in vivo. Science 213, 1264–1267 (1981)
Douglass, J. K. & Strausfeld, N. J. Anatomical organization of retinotopic motion—sensitive pathways in the optic lobes of flies. Microsc. Res. Tech. 62, 132–150 (2003)
Bausenwein, B., Dittrich, A. P. & Fischbach, K. F. The optic lobe of Drosophila melanogaster. II. Sorting of retinotopic pathways in the medulla. Cell Tissue Res. 267, 17–28 (1992)
Bausenwein, B. & Fischbach, K. F. Activity labeling patterns in the medulla of Drosophila melanogaster caused by motion stimuli. Cell Tissue Res. 270, 25–35 (1992)
Strausfeld, N. J. & Lee, J. K. Neuronal basis for parallel visual processing in the fly. Vis. Neurosci. 7, 13–33 (1991)
Clark, D. A., Bursztyn, L., Horowitz, M. A., Schnitzer, M. J. & Clandinin, T. R. Defining the computational structure of the motion detector in Drosophila. Neuron 70, 1165–1177 (2011)
Joesch, M., Schnell, B., Raghu, S. V., Reiff, D. F. & Borst, A. ON and OFF pathways in Drosophila motion vision. Nature 468, 300–304 (2010)
Rister, J. et al. Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron 56, 155–170 (2007)
Schnell, B., Raghu, S. V., Nern, A. & Borst, A. Columnar cells necessary for motion responses of wide-field visual interneurons in Drosophila. J. Comp. Physiol. A 198, 389–395 (2012)
Tuthill, J. C., Nern, A., Rubin, G. M. & Reiser, M. B. Contributions of the 12 neuron classes in the fly lamina to motion vision. Neuron 79, 128–140 (2013)
Douglass, J. K. & Strausfeld, N. J. Visual motion-detection circuits in flies: parallel direction-and non-direction-sensitive pathways between the medulla and lobula plate. J. Neurosci. 16, 4551–4562 (1996)
Maisak, M. S. et al. A directional tuning map of Drosophila elementary motion detectors. Nature http://dx.doi.org/10.1038/nature12320 (this issue)
Srinivasan, M. & Dvorak, D. Spatial processing of visual information in the movement-detecting pathway of the fly. J. Comp. Physiol. A 140, 1–23 (1980)
Haag, J. & Borst, A. Recurrent network interactions underlying flow-field selectivity of visual interneurons. J. Neurosci. 21, 5685–5692 (2001)
Gouwens, N. W. & Wilson, R. I. Signal propagation in Drosophila central neurons. J. Neurosci. 29, 6239–6249 (2009)
Ashmore, J. F. & Copenhagen, D. R. Different postsynaptic events in two types of retinal bipolar cell. Nature 288, 84–86 (1980)
Mizunami, M. Synaptic rectification model equivalent to the correlation-type movement detector. Biol. Cybern. 64, 1–6 (1990)
Oyster, C. W. & Barlow, H. B. Direction-selective units in rabbit retina: distribution of preferred directions. Science 155, 841–842 (1967)
Kim, I.-J., Zhang, Y., Yamagata, M., Meister, M. & Sanes, J. R. Molecular identification of a retinal cell type that responds to upward motion. Nature 452, 478–482 (2008)
Euler, T., Detwiler, P. B. & Denk, W. Directionally selective calcium signals in dendrites of starburst amacrine cells. Nature 418, 845–852 (2002)
Briggman, K. L., Helmstaedter, M. & Denk, W. Wiring specificity in the direction-selectivity circuit of the retina. Nature 471, 183–188 (2011)
Meinertzhagen, I. & Hanson, T. in The Development of Drosophila Melanogaster Vol. 2 (eds Bate, M. & Martinez Arias, A. ) 1363–1491 (Cold Spring Harbor Laboratory Press, 1993)
Blondel, V. D., Guillaume, J. L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008)
Eck, N. & Waltman, L. VOS: a new method for visualizing similarities between objects. Adv. Data Anal. 299–306 (2007)
Cardona, A. et al. TrakEM2 software for neural circuit reconstruction. PLoS ONE 7, e38011 (2012)
Scheffer, L., Karsh, B. & Vitaladevuni, S. Automated alignment of imperfect EM images for neural reconstruction. Preprint at http://arXiv.org/abs/1304.6034 (2013)
Canny, J. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. PAMI-8, 679–698 (1986)
Martin, D. R., Fowlkes, C. C. & Malik, J. Learning to detect natural image boundaries using local brightness, color, and texture cues. IEEE Trans. Pattern Anal. Mach. Intell. 26, 530–549 (2004)
Soille, P. Morphological Image Analysis: Principles and Applications 2nd edn, 316 (Springer-Verlag New York, 2003)
Dollar, P., Tu, Z. & Belongie, S. Supervised learning of edges and object boundaries. IEEE Comp. Soc. Conf. Comp. Vis. Pattern Rec. 2, 1964–1971 (2006)
Vitaladevuni, S., Mishchenko, Y., Genkin, A., Chklovskii, D. C. & Harris, K. M. Mitochondria detection in electron microscopy images. Workshop Microscopic Image Anal. Appl. Biol.. http://www.miaab.org/miaab-2008-papers/21-miaab-2008-abstract-05.pdf (2008)
Vincent, L. & Soille, P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 13, 583–598 (1991)
Mohanta, P. P., Mukherjee, D. P. & Acton, S. T. Agglomerative clustering for image segmentation. Int. Conf. Pattern Rec. 1, 664–667 (2002)
Vitaladevuni, S. N. & Basri, R. Co-clustering of image segments using convex optimization applied to EM neuronal reconstruction. IEEE Comp. Soc. Conf. Comp. Vis. Patt. Rec.. http://dx.doi.org/10.1109/CVPR.2010.5539901 2203–2210 (2010)
Kasai, H., Matsuzaki, M., Noguchi, J., Yasumatsu, N. & Nakahara, H. Structure–stability–function relationships of dendritic spines. Trends Neurosci. 26, 360–368 (2003)
Sato, M., Bitter, I., Bender, M. A., Kaufman, A. E. & Nakajima, M. TEASAR: Tree-structure extraction algorithm for accurate and robust skeletons. Eighth Pac. Conf. Comp. Graphics Appl.. http://dx.doi.org/10.1109/PCCGA.2000.883951 281–287, 449 (2000)
Acknowledgements
We acknowledge the technical support of all members of the Janelia FlyEM project and the Chklovskii group, past and present. We thank S. Laughlin for numerous discussions, and M. Reiser and N. Verma for commenting on the manuscript. We thank A. Borst, C. Desplan, C.-H. Lee, T. Clandinin and L. Zipursky for discussions and granting us access to their data before publication.
Author information
Authors and Affiliations
Contributions
D.B.C. and I.A.M. designed the research. Z.L., Sh.T. and R.D.F. prepared and imaged the sample. D.J.O., P.W., S.M.P., S.V. and W.T.K., under the guidance of D.B.C. and L.K.S., developed software for the reconstruction. Sh.T. annotated the micrographs, proofread the segmentation, and assembled the connectome, with the help of other proofreaders (Sa.T. K.B., L.-A.C., O.O., M.A.S., V.S. and C.S.), supervised by P.K.R. and J.A.H. A.N. and G.M.R. provided and A.N. analysed light microscopy images. L.K.S. and A.B. performed data analysis and T.Z. aided in visualization. A.B. and D.B.C. studied the motion detection circuit. A.B., D.B.C., I.A.M. and L.K.S. wrote the paper, with contributions from Sh.T. and A.N.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-6, Supplementary Methods, Supplementary Tables 1-3, a full legend to accompany the Supplementary Data file, the full video legend and Supplementary References. (PDF 6359 kb)
Supplementary Figure
This file contains the full version of Supplementary Figure 1 (see Supplementary Information file p.1 for full legend). (PDF 8306 kb)
Supplementary Table 1
This file contains the full version of Supplementary Table 1 (see Supplementary Information file p.16 for legend). (XLS 5614 kb)
Supplementary Data
This zipped file contains Supplementary Data 1 – see Supplementary Information file p.23 for legend). (ZIP 18850 kb)
Neuron reconstructions
This video shows the EM image stack of the medulla region of interest (Fig. 1c). Images from the stack are progressively removed (directed towards the deeper strata), and reconstructions of 379 neurons are added in succession, in randomly selected colors. The neurons are grouped into six classes, with text describing the class being added simultaneously with the neurons from each class: (1) Photoreceptor terminals, from the retina, and neurons from the lamina that innervate the reference column within the medulla. (2) Neurons receiving direct input from the retina and lamina neurons. (3) Neurons receiving direct input from the neurons in class (2). (4) Neurons that arborize in the reference column, but spread across multiple columns. (5) Tangential neurons, of which often only a fragment passing through the region of interest has been reconstructed. (6) Additional neurons, often of the same class as neurons in classes (1) – (5), but from adjacent columns. (MP4 29558 kb)
Rights and permissions
About this article
Cite this article
Takemura, Sy., Bharioke, A., Lu, Z. et al. A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181 (2013). https://doi.org/10.1038/nature12450
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12450
This article is cited by
-
Heterogeneity of synaptic connectivity in the fly visual system
Nature Communications (2024)
-
RoboEM: automated 3D flight tracing for synaptic-resolution connectomics
Nature Methods (2024)
-
How AI could lead to a better understanding of the brain
Nature (2023)
-
Different spectral sensitivities of ON- and OFF-motion pathways enhance the detection of approaching color objects in Drosophila
Nature Communications (2023)
-
Direction Selectivity of TmY Neurites in Drosophila
Neuroscience Bulletin (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.