Abstract
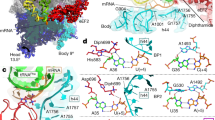
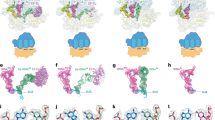
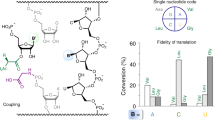
During normal translation, the binding of a release factor to one of the three stop codons (UGA, UAA or UAG) results in the termination of protein synthesis. However, modification of the initial uridine to a pseudouridine (Ψ) allows efficient recognition and read-through of these stop codons by a transfer RNA (tRNA), although it requires the formation of two normally forbidden purine–purine base pairs1. Here we determined the crystal structure at 3.1 Å resolution of the 30S ribosomal subunit in complex with the anticodon stem loop of tRNASer bound to the ΨAG stop codon in the A site. The ΨA base pair at the first position is accompanied by the formation of purine–purine base pairs at the second and third positions of the codon, which show an unusual Watson–Crick/Hoogsteen geometry. The structure shows a previously unsuspected ability of the ribosomal decoding centre to accommodate non-canonical base pairs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Karijolich, J. & Yu, Y.-T. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 474, 395–398 (2011)
Ogle, J. M. et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902 (2001)
Ogle, J. M., Murphy, F. V., Tarry, M. J. & Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002)
Schmeing, T. M. et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 (2009)
Voorhees, R. M., Schmeing, T. M., Kelley, A. C. & Ramakrishnan, V. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330, 835–838 (2010)
Pape, T., Wintermeyer, W. & Rodnina, M. V. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 17, 7490–7497 (1998)
Selmer, M. et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313, 1935–1942 (2006)
Leontis, N. B. & Westhof, E. Geometric nomenclature and classification of RNA base pairs. RNA 7, 499–512 (2001)
Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A. & Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153 (1998)
Nikolova, E. N. et al. Transient Hoogsteen base pairs in canonical duplex DNA. Nature 470, 498–502 (2011)
Davis, D. R., Veltri, C. A. & Nielsen, L. An RNA model system for investigation of pseudouridine stabilization of the codon-anticodon interaction in tRNALys, tRNAHis and tRNATyr. J. Biomol. Struct. Dyn. 15, 1121–1132 (1998)
Yarian, C. S. et al. Structural and functional roles of the N1- and N3-protons of Ψ at tRNA’s position 39. Nucleic Acids Res. 27, 3543–3549 (1999)
Tomita, K., Ueda, T. & Watanabe, K. The presence of pseudouridine in the anticodon alters the genetic code: a possible mechanism for assignment of the AAA lysine codon as asparagine in echinoderm mitochondria. Nucleic Acids Res. 27, 1683–1689 (1999)
Huang, C., Wu, G. & Yu, Y.-T. Inducing nonsense suppression by targeted pseudouridylation. Nature Protocols 7, 789–800 (2012)
Winn, M. D., Murshudov, G. N. & Papiz, M. Z. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374, 300–321 (2003)
Borel, F., Hartlein, M. & Leberman, R. In vivo overexpression and purification of Escherichia coli tRNAser. FEBS Lett. 324, 162–166 (1993)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
DeLano, W. L. The PyMOL Molecular Graphics System. http://www.pymol.org (2006)
Acknowledgements
We thank D. Hall and G. Winter for help and advice with data collection at beamline I04, Diamond Light Source; T. Tomizaki at beamline X06SA for help with data collection at the Swiss Light Source; and A. McCarthy at beamline ID14-4, ESRF, where screening and initial data collection were done. We thank M. Härtlein for the gift of an overproducing tRNASer clone, M. Torrent for advice on yeast tRNA abundance, and G. Murshudov for advice and help with data analysis and refinement. V.R. was supported by the UK Medical Research Council (grant U105184332), a Programme Grant and Senior Investigator Award from the Wellcome Trust, the Agouron Institute and the Louis-Jeantet Foundation. Y.-T.Y. was supported by a grant from the National Institutes of Health (GM104077), and by the University of Rochester CTSA award (UL1TR000042) from the National Center for Advancing Translational Sciences of the National Institutes of Health. I.S.F. was supported by a postdoctoral fellowship from the Fundacion Ramon Areces.
Author information
Authors and Affiliations
Contributions
I.S.F. carried out the crystallographic experiments and analysis and helped write the paper, G.W. did the in vitro translation assays, C.L.N. helped with crystallographic data collection, A.C.K. made the 30S subunits, 70S ribosome and tRNASer, and Y.-T.Y. and V.R. oversaw the project and helped write the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1 and Supplementary Figures 1-2. (PDF 1076 kb)
Rights and permissions
About this article
Cite this article
Fernández, I., Ng, C., Kelley, A. et al. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature 500, 107–110 (2013). https://doi.org/10.1038/nature12302
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12302
This article is cited by
-
Deciphering glioma epitranscriptome: focus on RNA modifications
Oncogene (2023)
-
Quantitative sequencing using BID-seq uncovers abundant pseudouridines in mammalian mRNA at base resolution
Nature Biotechnology (2023)
-
The importance of pseudouridylation: human disorders related to the fifth nucleoside
Biologia Futura (2023)
-
snoRNAs: functions and mechanisms in biological processes, and roles in tumor pathophysiology
Cell Death Discovery (2022)
-
Epitranscriptomic dynamics in brain development and disease
Molecular Psychiatry (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.