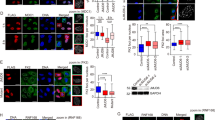
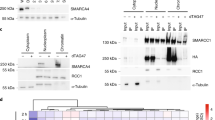
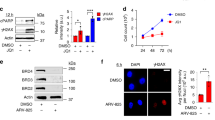
Abstract
DNA damage activates a signalling network that blocks cell-cycle progression, recruits DNA repair factors and/or triggers senescence or programmed cell death1. Alterations in chromatin structure are implicated in the initiation and propagation of the DNA damage response2. Here we further investigate the role of chromatin structure in the DNA damage response by monitoring ionizing-radiation-induced signalling and response events with a high-content multiplex RNA-mediated interference screen of chromatin-modifying and -interacting genes. We discover that an isoform of Brd4, a bromodomain and extra-terminal (BET) family member, functions as an endogenous inhibitor of DNA damage response signalling by recruiting the condensin II chromatin remodelling complex to acetylated histones through bromodomain interactions. Loss of this isoform results in relaxed chromatin structure, rapid cell-cycle checkpoint recovery and enhanced survival after irradiation, whereas functional gain of this isoform compacted chromatin, attenuated DNA damage response signalling and enhanced radiation-induced lethality. These data implicate Brd4, previously known for its role in transcriptional control, as an insulator of chromatin that can modulate the signalling response to DNA damage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Jackson, S. P. & Bartek, J. The DNA-damage response in human biology and disease. Nature 461, 1071–1078 (2009)
Misteli, T. & Soutoglou, E. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nature Rev. Mol. Cell Biol. 10, 243–254 (2009)
Gorgoulis, V. G. et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 (2005)
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005)
Kastan, M. B. & Bartek, J. Cell-cycle checkpoints and cancer. Nature 432, 316–323 (2004)
Polo, S. E. & Jackson, S. P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25, 409–433 (2011)
Moffat, J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006)
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006)
Rahman, S. et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol. Cell. Biol. 31, 2641–2652 (2011)
Yang, Z. et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 19, 535–545 (2005)
Jang, M. K. et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19, 523–534 (2005)
Murga, M. et al. Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 178, 1101–1108 (2007)
Ziv, Y. et al. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nature Cell Biol. 8, 870–876 (2006)
Cowell, I. G. et al. γH2AX foci form preferentially in euchromatin after ionising-radiation. PLoS ONE 2, e1057 (2007)
Kim, J. A., Kruhlak, M., Dotiwala, F., Nussenzweig, A. & Haber, J. E. Heterochromatin is refractory to γ-H2AX modification in yeast and mammals. J. Cell Biol. 178, 209–218 (2007)
Filippakopoulos, P. et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 149, 214–231 (2012)
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010)
Bradner, J. E. et al. Chemical phylogenetics of histone deacetylases. Nature Chem. Biol. 6, 238–243 (2010)
Wu, N. & Yu, H. The Smc complexes in DNA damage response. Cell Biosci. 2, 5 (2012)
Lee, H.-S., Park, J.-H., Kim, S.-J., Kwon, S.-J. & Kwon, J. A cooperative activation loop among SWI/SNF, γ-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 29, 1434–1445 (2010)
Smyth, G. K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, Article3 (2004)
Carey, M. & Smale, S. T. Micrococcal nuclease-Southern blot assay: I. MNase and restriction digestions. CSH Protoc. 2007, http://dx.doi.org/10.1101/pdb.prot4890 (2007)
Acknowledgements
We thank H. Le, T.R. Jones and M. Vokes for assistance with screening and image analysis. We thank C. Whittaker, S. Hoersch and M. Moran for computing and data analysis assistance; C. Reinhardt, C. Ellson and A. Gardino for manuscript editing; and P. Filippakopoulos and S. Knapp for discussions. This work was partially supported by the Koch Institute and Center for Environmental Health Sciences National Institutes of Health Core Grants P30-CA14051 and ES-002109; and by grants R01-ES15339, 1-U54-CA112967-04 and R21-NS063917; a SPARC grant to M.B.Y.; and a Holman Pathway Research Resident Seed Grant, American Society for Radiation Oncology Junior Faculty Career Research Training Award, Klarman Scholar, Koch Institute Clinical Investigator Award, and Burroughs Wellcome Career Award for Medical Scientists to S.R.F.
Author information
Authors and Affiliations
Contributions
S.R.F. and M.B.Y. designed the study, supervised the experiments, analysed the data and wrote the manuscript. D.E.R., W.C.H. and D.M.S. were involved in the design and preparation of the lentiviral shRNA library. S.R.F., M.E.P. and E.B. performed the image-based high-content screen and initial analysis. A.E.C. aided in digital image analysis. S.R.F., Q.H., S.M.C., F.C.L., I.G.C., M.J.L., A.F., R.H., B.A.G., G.C.C. and A.M. performed biochemical, cell biological and molecular biological experiments. B.D.B., A.M.D. and F.M.W. performed mass spectrometry experiments and analysis. J.R. performed bioinformatics analysis. J.E.B. contributed JQ1 compounds and cell lines. S.R.F. and M.B.Y. designed and supervised the experiments. C.C.C., J.E.B. and F.M.W. contributed to the intellectual development of the study and technical writing of the manuscript. All authors contributed to editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-14. (PDF 1573 kb)
Supplementary Table 1
This file contains γH2AX and nuclear features from high-content shRNA screen. U2OS cells were screened in 384-well plate format using shRNA directed against the indicated gene symbols by the method as outlined in Figure 1 and in the Online Methods. Listed are measurements for several features of identified nuclei and γH2AX foci for each hairpin at all timepoints. (XLS 1579 kb)
Supplementary Table 2
This file contains modulators of γH2AX foci number, intensity and size. List of the top quartile of genes ranked by increasing γH2AX foci number per nucleus, area, and integrated fluorescence intensity 1 and 6 hours following 10 Gy IR. Genes that appear shaded in green scored in the top quartile at both the 1 and 6 hour time points. (XLS 44 kb)
Supplementary Table 3
This file contains Brd4 isoform B interacting proteins. A list of peptides, associated genes, and MASCOT scores identified as Brd4 isoform B interactors by mass spectrometry from U2OS cell immunoprecipitates from two independent experiments. Genes that appear shaded in green were identified in experimental replicates 1 and 2. (XLS 33 kb)
Supplementary Table 4
This file contains Brd4 isoform interactions with SMC proteins. A list of protein scores, unique and total number of peptides, peptide sequences and MASCOT scores for peptides identified by mass spectrometry of Commassie Brilliant Blue stained gel regions as indicated in Supplementary Figure 7. (XLS 41 kb)
Rights and permissions
About this article
Cite this article
Floyd, S., Pacold, M., Huang, Q. et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature 498, 246–250 (2013). https://doi.org/10.1038/nature12147
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12147
This article is cited by
-
High-sensitive nascent transcript sequencing reveals BRD4-specific control of widespread enhancer and target gene transcription
Nature Communications (2023)
-
Methylation of BRD4 by PRMT1 regulates BRD4 phosphorylation and promotes ovarian cancer invasion
Cell Death & Disease (2023)
-
Inhibition of demethylase by IOX1 modulates chromatin accessibility to enhance NSCLC radiation sensitivity through attenuated PIF1
Cell Death & Disease (2023)
-
Ablative radiotherapy improves survival but does not cure autochthonous mouse models of prostate and colorectal cancer
Communications Medicine (2023)
-
The consequences of viral infection on host DNA damage response: a focus on SARS-CoVs
Journal of Genetic Engineering and Biotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.