Abstract
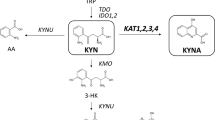
Inhibition of kynurenine 3-monooxygenase (KMO), an enzyme in the eukaryotic tryptophan catabolic pathway (that is, kynurenine pathway), leads to amelioration of Huntington’s-disease-relevant phenotypes in yeast, fruitfly and mouse models1,2,3,4,5, as well as in a mouse model of Alzheimer’s disease3. KMO is a flavin adenine dinucleotide (FAD)-dependent monooxygenase and is located in the outer mitochondrial membrane where it converts l-kynurenine to 3-hydroxykynurenine. Perturbations in the levels of kynurenine pathway metabolites have been linked to the pathogenesis of a spectrum of brain disorders6, as well as cancer7,8 and several peripheral inflammatory conditions9. Despite the importance of KMO as a target for neurodegenerative disease, the molecular basis of KMO inhibition by available lead compounds has remained unknown. Here we report the first crystal structure of Saccharomyces cerevisiae KMO, in the free form and in complex with the tight-binding inhibitor UPF 648. UPF 648 binds close to the FAD cofactor and perturbs the local active-site structure, preventing productive binding of the substrate l-kynurenine. Functional assays and targeted mutagenesis reveal that the active-site architecture and UPF 648 binding are essentially identical in human KMO, validating the yeast KMO–UPF 648 structure as a template for structure-based drug design. This will inform the search for new KMO inhibitors that are able to cross the blood–brain barrier in targeted therapies against neurodegenerative diseases such as Huntington’s, Alzheimer’s and Parkinson’s diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Giorgini, F. et al. Histone deacetylase inhibition modulates kynurenine pathway activation in yeast, microglia, and mice expressing a mutant huntingtin fragment. J. Biol. Chem. 283, 7390–7400 (2008)
Giorgini, F., Guidetti, P., Nguyen, Q., Bennett, S. C. & Muchowski, P. J. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nature Genet. 37, 526–531 (2005)
Zwilling, D. et al. Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell 145, 863–874 (2011)
Campesan, S. et al. The kynurenine pathway modulates neurodegeneration in a Drosophila model of Huntington’s disease. Curr. Biol. 21, 961–966 (2011)
Green, E. W. et al. Drosophila eye color mutants as therapeutic tools for Huntington disease. Fly 6, 117–120 (2012)
Schwarcz, R., Bruno, J. P., Muchowski, P. J. & Wu, H.-Q. Kynurenines in the mammalian brain: when physiology meets pathology. Nature Rev. Neurosci. 13, 465–477 (2012)
Platten, M., Litzenburger, U. & Wick, W. The aryl hydrocarbon receptor in tumor immunity. Oncoimmunology 1, 396–397 (2012)
Liu, X., Newton, R. C., Friedman, S. M. & Scherle, P. A. Indoleamine 2,3-dioxygenase, an emerging target for anti-cancer therapy. Curr. Cancer Drug Targets 9, 938–952 (2009)
Filippini, P. et al. Emerging concepts on inhibitors of indoleamine 2,3-dioxygenase in rheumatic diseases. Curr. Med. Chem. 19, 5381–5393 (2012)
Stone, T. W. & Perkins, M. N. Quinolinic acid: a potent endogenous excitant at amino acid receptors in CNS. Eur. J. Pharmacol. 72, 411–412 (1981)
Schwarcz, R., Whetsell, W. O., Jr & Mangano, R. M. Quinolinic acid: an endogenous metabolite that produces axon-sparing lesions in rat brain. Science 219, 316–318 (1983)
Okuda, S., Nishiyama, N., Saito, H. & Katsuki, H. Hydrogen peroxide-mediated neuronal cell death induced by an endogenous neurotoxin, 3-hydroxykynurenine. Proc. Natl Acad. Sci. USA 93, 12553–12558 (1996)
Copeland, C. S., Neale, S. A. & Salt, T. E. Actions of xanthurenic acid, a putative endogenous group II metabotropic glutamate receptor agonist, on sensory transmission in the thalamus. Neuropharmacology 66, 133–142 (2013)
Fazio, F. et al. Cinnabarinic acid, an endogenous metabolite of the kynurenine pathway, activates type 4 metabotropic glutamate receptors. Mol. Pharmacol. 81, 643–656 (2012)
Guillemin, G. J. et al. Characterization of the kynurenine pathway in human neurons. J. Neurosci. 27, 12884–12892 (2007)
Guillemin, G. J., Smith, D. G., Smythe, G. A., Armati, P. J. & Brew, B. J. Expression of the kynurenine pathway enzymes in human microglia and macrophages. Adv. Exp. Med. Biol. 527, 105–112 (2003)
Cozzi, A., Carpenedo, R. & Moroni, F. Kynurenine hydroxylase inhibitors reduce ischemic brain damage: studies with (m-nitrobenzoyl)-alanine (mNBA) and 3,4-dimethoxy-[-N-4-(nitrophenyl)thiazol-2yl]-benzenesulfonamide (Ro 61–8048) in models of focal or global brain ischemia. J. Cereb. Blood Flow Metab. 19, 771–777 (1999)
Moroni, F. et al. Studies on the neuroprotective action of kynurenine mono-oxygenase inhibitors in post-ischemic brain damage. Adv. Exp. Med. Biol. 527, 127–136 (2003)
Richter, A. & Hamann, M. The kynurenine 3-hydroxylase inhibitor Ro 61–8048 improves dystonia in a genetic model of paroxysmal dyskinesia. Eur. J. Pharmacol. 478, 47–52 (2003)
Samadi, P. et al. Effect of kynurenine 3-hydroxylase inhibition on the dyskinetic and antiparkinsonian responses to levodopa in Parkinsonian monkeys. Mov. Disord. 20, 792–802 (2005)
Clark, C. J. et al. Prolonged survival of a murine model of cerebral malaria by kynurenine pathway inhibition. Infect. Immun. 73, 5249–5251 (2005)
Reinhart, P. H. & Kelly, J. W. Treating the periphery to ameliorate neurodegenerative diseases. Cell 145, 813–814 (2011)
Sapko, M. T. et al. Endogenous kynurenate controls the vulnerability of striatal neurons to quinolinate: implications for Huntingtons’s disease. Exp. Neurol. 197, 31–40 (2006)
Ceresoli-Borroni, G., Guidetti, P., Amori, L., Pellicciari, R. & Schwarcz, R. Perinatal kynurenine 3-hydroxylase inhibition in rodents: pathophysiological implications. J. Neurosci. Res. 85, 845–854 (2007)
Uemura, T. & Hirai, K. l-kynurenine 3-monooxygenase from mitochondrial outer membrane of pig liver: purification, some properties, and monoclonal antibodies directed to the enzyme. J. Biochem. 123, 253–262 (1998)
Palfey, B. A. & McDonald, C. A. Control of catalysis in flavin-dependent monooxygenases. Arch. Biochem. Biophys. 493, 26–36 (2010)
van Berkel, W. J. H., Kamerbeek, N. M. & Fraaije, M. W. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124, 670–689 (2006)
McCulloch, K. M., Mukherjee, T., Begley, T. P. & Ealick, S. E. Structure of the PLP degradative enzyme 2-methyl-3- hydroxypyridine-5-carboxylic acid oxygenase from Mesorhizobium loti MAFF303099 and its mechanistic implications. Biochemistry 48, 4139–4149 (2009)
Ortiz-Maldonado, M., Ballou, D. P. & Massey, V. A rate-limiting conformational change of the flavin in p-hydroxybenzoate hydroxylase is necessary for ligand exchange and catalysis: studies with 8-mercapto- and 8-hydroxy-flavins. Biochemistry 40, 1091–1101 (2001)
Fukui, S., Schwarcz, R., Rapoport, S. I., Takada, Y. & Smith, Q. R. Blood–brain barrier transport of kynurenines: implications for brain synthesis and metabolism. J. Neurochem. 56, 2007–2017 (1991)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
McCoy, A. J., Storoni, L. C. & Read, R. J. Simple algorithm for a maximum-likelihood SAD function. Acta Crystallogr. D 60, 1220–1228 (2004)
Terwilliger, T. C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D 64, 61–69 (2008)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005)
Cornell, W. D. et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 (1995)
Jakalian, A., Jack, D. B. & Bayly, C. I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 23, 1623–1641 (2002)
Acknowledgements
We thank R. Schwarcz for supplying UPF 648. We also thank E. McKenzie for expressing human KMO. We also thank Diamond Light Source for access to MX beamlines.
Author information
Authors and Affiliations
Contributions
N.S.S., F.G., D.L. and T.F.O. initiated the project, designed experiments, analysed data and wrote the manuscript; M.A. cloned purified and crystallized proteins and performed biochemical assays; C.L. crystallized proteins, collected and processed diffraction data; D.J.H. developed and analysed some of the biochemical assays; P.L. performed molecular modelling of l-KYN binding.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-5 and Supplementary Tables 1-3. (PDF 1183 kb)
Rights and permissions
About this article
Cite this article
Amaral, M., Levy, C., Heyes, D. et al. Structural basis of kynurenine 3-monooxygenase inhibition. Nature 496, 382–385 (2013). https://doi.org/10.1038/nature12039
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12039
This article is cited by
-
The kynurenine pathway and bipolar disorder: intersection of the monoaminergic and glutamatergic systems and immune response
Molecular Psychiatry (2021)
-
Functional and structural characterization of a flavoprotein monooxygenase essential for biogenesis of tryptophylquinone cofactor
Nature Communications (2021)
-
Full-length in meso structure and mechanism of rat kynurenine 3-monooxygenase inhibition
Communications Biology (2021)
-
Redox signaling and Alzheimer’s disease: from pathomechanism insights to biomarker discovery and therapy strategy
Biomarker Research (2020)
-
Targeting tryptophan catabolic kynurenine pathway enhances antitumor immunity and cytotoxicity in multiple myeloma
Leukemia (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.