Abstract
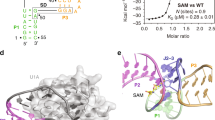
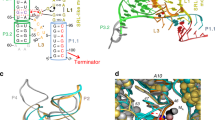
Structures of riboswitch receptor domains bound to their effector have shown how messenger RNAs recognize diverse small molecules, but mechanistic details linking the structures to the regulation of gene expression remain elusive1,2. To address this, here we solve crystal structures of two different classes of cobalamin (vitamin B12)-binding riboswitches that include the structural switch of the downstream regulatory domain. These classes share a common cobalamin-binding core, but use distinct peripheral extensions to recognize different B12 derivatives. In each case, recognition is accomplished through shape complementarity between the RNA and cobalamin, with relatively few hydrogen bonding interactions that typically govern RNA–small molecule recognition. We show that a composite cobalamin–RNA scaffold stabilizes an unusual long-range intramolecular kissing-loop interaction that controls mRNA expression. This is the first, to our knowledge, riboswitch crystal structure detailing how the receptor and regulatory domains communicate in a ligand-dependent fashion to regulate mRNA expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Garst, A. D., Edwards, A. L. & Batey, R. T. Riboswitches: structures and mechanisms. Cold Spring Harb. Perspect. Biol. 3, a003533 (2011)
Breaker, R. R. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 4, a003566 (2012)
Lundrigan, M. D., Koster, W. & Kadner, R. J. Transcribed sequences of the Escherichia coli btuB gene control its expression and regulation by vitamin B12 . Proc. Natl Acad. Sci. USA 88, 1479–1483 (1991)
Franklund, C. V. & Kadner, R. J. Multiple transcribed elements control expression of the Escherichia coli btuB gene. J. Bacteriol. 179, 4039–4042 (1997)
Nahvi, A., Barrick, J. E. & Breaker, R. R. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 32, 143–150 (2004)
Nahvi, A. et al. Genetic control by a metabolite binding mRNA. Chem. Biol. 9, 1043 (2002)
Rodionov, D. A., Vitreschak, A. G., Mironov, A. A. & Gelfand, M. S. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278, 41148–41159 (2003)
Vitreschak, A. G., Rodionov, D. A., Mironov, A. A. & Gelfand, M. S. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA 9, 1084–1097 (2003)
Barrick, J. E. & Breaker, R. R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8, R239 (2007)
Gardner, P. P. et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 37, D136–D140 (2009)
Fox, K. et al. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc. Natl Acad. Sci. USA 106, 4435–4440 (2009)
Wilkinson, K. A., Merino, E. J. & Weeks, K. M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nature Protocols 1, 1610–1616 (2006)
Lundrigan, M. D. & Kadner, R. J. Altered cobalamin metabolism in Escherichia coli btuR mutants affects btuB gene regulation. J. Bacteriol. 171, 154–161 (1989)
Weinberg, Z. et al. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 11, R31 (2010)
Taylor, R. T., Smucker, L., Hanna, M. L. & Gill, J. Aerobic photolysis of alkylcobalamins: quantum yields and light-action spectra. Arch. Biochem. Biophys. 156, 521–533 (1973)
Adams, P. L., Stahley, M. R., Kosek, A. B., Wang, J. & Strobel, S. A. Crystal structure of a self-splicing group I intron with both exons. Nature 430, 45–50 (2004)
Jaeger, L., Verzemnieks, E. J. & Geary, C. The UA_handle: a versatile submotif in stable RNA architectures. Nucleic Acids Res. 37, 215–230 (2009)
Nagaswamy, U. & Fox, G. E. Frequent occurrence of the T-loop RNA folding motif in ribosomal RNAs. RNA 8, 1112–1119 (2002)
Krasilnikov, A. S. & Mondragon, A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA 9, 640–643 (2003)
Serganov, A., Huang, L. & Patel, D. J. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 458, 233–237 (2009)
Sussman, D., Nix, J. C. & Wilson, C. The structural basis for molecular recognition by the vitamin B12 RNA aptamer. Nature Struct. Biol. 7, 53–57 (2000)
Ortiz-Guerrero, J. M., Polanco, M. C., Murillo, F. J., Padmanabhan, S. & Elias-Arnanz, M. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc. Natl Acad. Sci. USA 108, 7565–7570 (2011)
Vicens, Q. & Cech, T. R. Atomic level architecture of group I introns revealed. Trends Biochem. Sci. 31, 41–51 (2006)
Melnikov, S. et al. One core, two shells: bacterial and eukaryotic ribosomes. Nature Struct. Mol. Biol. 19, 560–567 (2012)
Xin, Y., Laing, C., Leontis, N. B. & Schlick, T. Annotation of tertiary interactions in RNA structures reveals variations and correlations. RNA 14, 2465–2477 (2008)
Butcher, S. E. & Pyle, A. M. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 44, 1302–1311 (2011)
Gregorian, R. S., Jr & Crothers, D. M. Determinants of RNA hairpin loop-loop complex stability. J. Mol. Biol. 248, 968–984 (1995)
Pyle, A. M. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 7, 679–690 (2002)
Reyes, F. E., Garst, A. D. & Batey, R. T. Strategies in RNA crystallography. Methods Enzymol. 469, 119–139 (2009)
Edwards, A. L., Garst, A. D. & Batey, R. T. Determining structures of RNA aptamers and riboswitches by X-ray crystallography. Methods Mol. Biol. 535, 135–163 (2009)
Milligan, J. F. & Uhlenbeck, O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 180, 51–62 (1989)
Gilbert, S. D. & Batey, R. T. Monitoring RNA–ligand interactions using isothermal titration calorimetry. Methods Mol. Biol. 540, 97–114 (2009)
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006)
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D 67, 293–302 (2011)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D 60, 2210–2221 (2004)
Keating, K. S. & Pyle, A. M. Semiautomated model building for RNA crystallography using a directed rotameric approach. Proc. Natl Acad. Sci. USA 107, 8177–8182 (2010)
Ferré-D’Amaré, A. R. Use of the spliceosomal protein U1A to facilitate crystallization and structure determination of complex RNAs. Methods 52, 159–167 (2010)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Acknowledgements
This work was supported by grants from the National Institutes of Health (GM073850 and 1S10RR026516) to R.T.B. and by a Colorado Diversity Initiative Fellowship and NIH Ruth L. Kirschstein fellowship (F32GM095121) to J.E.J. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under contract no. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Contributions
F.E.R. discovered the specificity of the AqCbl class and performed all aspects of the crystallography with assistance from J.T.P.; J.E.J. performed all biochemical experiments and fully characterized the specificities of the cobalamin family; J.T.P. obtained all in vivo data; and all authors contributed to the analysis of the data and the writing of this paper.
Corresponding authors
Ethics declarations
Competing interests
R.T.B. is a member of the Scientific Advisory Board of BioRelix, a company pursuing the development of antimicrobials that target riboswitches.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-12, Supplementary Tables 1-3 and additional references. (PDF 27942 kb)
Rights and permissions
About this article
Cite this article
Johnson Jr, J., Reyes, F., Polaski, J. et al. B12 cofactors directly stabilize an mRNA regulatory switch. Nature 492, 133–137 (2012). https://doi.org/10.1038/nature11607
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11607
This article is cited by
-
The Joint Anaerobic Denitrification Performance of Klebsiella sp. and Enterobacter hormaechei Using Two Carbon Substrates With and Without the Presence of Heavy Metals
Water, Air, & Soil Pollution (2022)
-
A multicolor riboswitch-based platform for imaging of RNA in live mammalian cells
Nature Chemical Biology (2018)
-
Microbial production of vitamin B12: a review and future perspectives
Microbial Cell Factories (2017)
-
Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation
Nature Communications (2017)
-
Effects of heavy metals on aerobic denitrification by strain Pseudomonas stutzeri PCN-1
Applied Microbiology and Biotechnology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.