Abstract
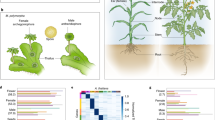
Animal and plant development starts with a constituting phase called embryogenesis, which evolved independently in both lineages1. Comparative anatomy of vertebrate development—based on the Meckel-Serrès law2 and von Baer’s laws of embryology3 from the early nineteenth century—shows that embryos from various taxa appear different in early stages, converge to a similar form during mid-embryogenesis, and again diverge in later stages. This morphogenetic series is known as the embryonic ‘hourglass’4,5, and its bottleneck of high conservation in mid-embryogenesis is referred to as the phylotypic stage6. Recent analyses in zebrafish and Drosophila embryos provided convincing molecular support for the hourglass model, because during the phylotypic stage the transcriptome was dominated by ancient genes7 and global gene expression profiles were reported to be most conserved8. Although extensively explored in animals, an embryonic hourglass has not been reported in plants, which represent the second major kingdom in the tree of life that evolved embryogenesis. Here we provide phylotranscriptomic evidence for a molecular embryonic hourglass in Arabidopsis thaliana, using two complementary approaches. This is particularly significant because the possible absence of an hourglass based on morphological features in plants suggests that morphological and molecular patterns might be uncoupled. Together with the reported developmental hourglass patterns in animals, these findings indicate convergent evolution of the molecular hourglass and a conserved logic of embryogenesis across kingdoms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Meyerowitz, E. M. Plants compared to animals: the broadest comparative study of development. Science 295, 1482–1485 (2002)
Meckel, J. F. Beyträge zur vergleichenden Anatomie (Reclam, Leipzig, 1811)
von Baer, K. E. Über Entwicklungsgeschichte der Thiere: Beobachtung und Reflexion (Gebrüder Bornträger, Königsberg, 1828)
Duboule, D. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev. Suppl. 1994135–142 (1994)
Raff, R. A. The Shape of Life: Genes, Development and the Evolution of Animal Form (Univ. Chicago Press, 1996)
Sander, K. in Development and Evolution (eds Goodwin, B. C., Holder, N. & Wylie, C. C. ) 137–159 (Cambridge Univ. Press, 1983)
Domazet-Lošo, T. & Tautz, D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468, 815–818 (2010)
Kalinka, A. T. et al. Gene expression divergence recapitulates the developmental hourglass model. Nature 468, 811–814 (2010)
De Smet, I., Lau, S., Mayer, U. & Jürgens, G. Embryogenesis — the humble beginnings of plant life. Plant J. 61, 959–970 (2010)
Peris, C. I., Rademacher, E. H. & Weijers, D. Green beginnings — pattern formation in the early plant embryo. Curr. Top. Dev. Biol. 91, 1–27 (2010)
Xiang, D. et al. Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol. 156, 346–356 (2011)
Domazet-Lošo, T., Brajković, J. & Tautz, D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 23, 533–539 (2007)
Levin, M., Hashimshony, T., Wagner, F. & Yanai, I. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev. Cell 22, 1101–1108 (2012)
Koch, M. A., Haubold, B. & Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17, 1483–1498 (2000)
Arakaki, M. et al. Contemporaneous and recent radiations of the world's major succulent plant lineages. Proc. Natl Acad. Sci. USA 108, 8379–8384 (2011)
Koch, M. A. & Kiefer, M. Genome evolution among cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species — Capsella rubella, Arabidopsis lyrata subsp. petraea, and A. thaliana.. Am. J. Bot. 92, 761–767 (2005)
Oh, D.-H. et al. Genome structures and halophyte-specific gene expression of the extremophile Thellungiella parvula in comparison with Thellungiella salsuginea (Thellungiella halophila) and Arabidopsis. Plant Physiol. 154, 1040–1052 (2010)
Zuber, H. et al. The seed composition of Arabidopsis mutants for the group 3 sulfate transporters indicates a role in sulfate translocation within developing seeds. Plant Physiol. 154, 913–926 (2010)
Le, B. H. et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl Acad. Sci. USA 107, 8063–8070 (2010)
Nodine, M. D. & Bartel, D. P. Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature 482, 94–97 (2012)
Slack, J. M., Holland, P. W. & Graham, C. F. The zootype and the phylotypic stage. Nature 361, 490–492 (1993)
Lau, S., Slane, D., Herud, O., Kong, J. & Jürgens, G. Early embryogenesis in flowering plants: setting up the basic body pattern. Annu. Rev. Plant Biol. 63, 483–506 (2012)
Park, S. & Harada, J. J. Arabidopsis embryogenesis. Methods Mol. Biol. 427, 3–16 (2008)
Irie, N. & Kuratani, S. Comparative transcriptome analysis reveals vertebrate phylotypic period during organogenesis. Nature Commun 2, 248 (2011)
Irie, N. & Sehara-Fujisawa, A. The vertebrate phylotypic stage and an early bilaterian-related stage in mouse embryogenesis defined by genomic information. BMC Biol. 5, 1 (2007)
Katoh, K., Kuma, K., Toh, H. & Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005)
Suyama, M., Torrents, D. & Bork, P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006)
Thornton, K. Libsequence: a C++ class library for evolutionary genetic analysis. Bioinformatics 19, 2325–2327 (2003)
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–1186 (2012)
Wu, Z. et al. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99, 909–917 (2004)
Carbon, S. et al. AmiGO: online access to ontology and annotation data. Bioinformatics 25, 288–289 (2009)
Acknowledgements
We are grateful to S. Abel, L. I. A. Calderón Villalobos, C. Delker, T. Greb, D. Grubb, J. J. Harada, D. Jackson, R. Paxton, A. Soro and C. Wasternack for discussions, to S. Neumann for support with the IPB-cluster, and the 'Exzellenznetzwerk für Biowissenschaften' of the Federal State of Sachsen-Anhalt, Germany, for financial support of M.Q.
Author information
Authors and Affiliations
Contributions
M.Q. conceived the study. M.Q. and I.G. supervised the project. H.-G.D., A.G., K.K.U. and M.B. analysed the data. M.Q., H.-G.D., A.G., K.K.U., M.B. and I.G. interpreted the results. M.Q. and I.G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Supplementary Figures 1-17, Supplementary Tables 1, 5 and 6 (see separate files for Supplementary Tables 2, 3 and 4). (PDF 1654 kb)
Supplementary Table 2
This file contains the phylostrata information and Ka/Ks raw data for each A. thaliana gene for which we identified an ortholog in A. lyrata. (XLS 5974 kb)
Supplementary Table 3
This file contains the phylostrata information and Ka/Ks raw data for each A. thaliana gene for which we identified an ortholog in A. lyrata. (XLS 10378 kb)
Supplementary Table 4
This file contains the phylostrata information and Ka/Ks raw data for each A. thaliana gene for which we identified an ortholog in A. lyrata. (XLS 10352 kb)
Rights and permissions
About this article
Cite this article
Quint, M., Drost, HG., Gabel, A. et al. A transcriptomic hourglass in plant embryogenesis. Nature 490, 98–101 (2012). https://doi.org/10.1038/nature11394
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11394
This article is cited by
-
Macroevolutionary dynamics of gene family gain and loss along multicellular eukaryotic lineages
Nature Communications (2024)
-
How ancient genes form animal body plans
Nature Reviews Genetics (2024)
-
Dynamic chromatin regulatory programs during embryogenesis of hexaploid wheat
Genome Biology (2023)
-
Conserved, divergent and heterochronic gene expression during Brachypodium and Arabidopsis embryo development
Plant Reproduction (2021)
-
Evolutionary transcriptomics of metazoan biphasic life cycle supports a single intercalation origin of metazoan larvae
Nature Ecology & Evolution (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.