Abstract
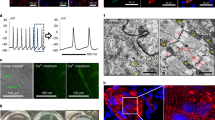
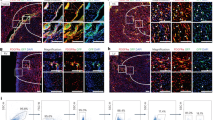
The adult mammalian heart possesses little regenerative potential following injury. Fibrosis due to activation of cardiac fibroblasts impedes cardiac regeneration and contributes to loss of contractile function, pathological remodelling and susceptibility to arrhythmias. Cardiac fibroblasts account for a majority of cells in the heart and represent a potential cellular source for restoration of cardiac function following injury through phenotypic reprogramming to a myocardial cell fate. Here we show that four transcription factors, GATA4, HAND2, MEF2C and TBX5, can cooperatively reprogram adult mouse tail-tip and cardiac fibroblasts into beating cardiac-like myocytes in vitro. Forced expression of these factors in dividing non-cardiomyocytes in mice reprograms these cells into functional cardiac-like myocytes, improves cardiac function and reduces adverse ventricular remodelling following myocardial infarction. Our results suggest a strategy for cardiac repair through reprogramming fibroblasts resident in the heart with cardiogenic transcription factors or other molecules.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lopez, A. D., Mathers, C. D., Ezzati, M., Jamison, D. T. & Murray, C. J. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet 367, 1747–1757 (2006)
Porrello, E. R. et al. Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 (2011)
Yacoub, M., Suzuki, K. & Rosenthal, N. The future of regenerative therapy in patients with chronic heart failure. Nat. Clin. Pract. Cardiovasc. Med. 3 (Suppl. 1). S133–S135 (2006)
Menasche, P. Cell-based therapy for heart disease: a clinically oriented perspective. Mol. Ther. 17, 758–766 (2009)
Hoover-Plow, J. & Gong, Y. Challenges for heart disease stem cell therapy. Vasc. Health Risk Manag. 8, 99–113 (2012)
Perin, E. C. et al. Effect of transendocardial delivery of autologous bone marrow mononuclear cells on functional capacity, left ventricular function, and perfusion in chronic heart failure: the FOCUS-CCTRN trial. J. Am. Med. Assoc. (2012)
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006)
Yu, J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007)
Tapscott, S. J. et al. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405–411 (1988)
Wang, Z., Wang, D. Z., Pipes, G. C. & Olson, E. N. Myocardin is a master regulator of smooth muscle gene expression. Proc. Natl Acad. Sci. USA 100, 7129–7134 (2003)
Vierbuchen, T. et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010)
Caiazzo, M. et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227 (2011)
Jugdutt, B. I. Ventricular remodeling after infarction and the extracellular collagen matrix: when is enough enough? Circulation 108, 1395–1403 (2003)
Olson, E. N. Gene regulatory networks in the evolution and development of the heart. Science 313, 1922–1927 (2006)
Bondue, A. & Blanpain, C. Mesp1: a key regulator of cardiovascular lineage commitment. Circ. Res. 107, 1414–1427 (2010)
Ieda, M. et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 (2010)
Zang, M. X., Li, Y., Wang, H., Wang, J. B. & Jia, H. T. Cooperative interaction between the basic helix-loop-helix transcription factor dHAND and myocyte enhancer factor 2C regulates myocardial gene expression. J. Biol. Chem. 279, 54258–54263 (2004)
Dai, Y. S., Cserjesi, P., Markham, B. E. & Molkentin, J. D. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J. Biol. Chem. 277, 24390–24398 (2002)
Garg, V. et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424, 443–447 (2003)
Ghosh, T. K. et al. Physical interaction between TBX5 and MEF2C is required for early heart development. Mol. Cell. Biol. 29, 2205–2218 (2009)
Warren, L. et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, 618–630 (2010)
Porter, K. E. & Turner, N. A. Cardiac fibroblasts: at the heart of myocardial remodeling. Pharmacol. Ther. 123, 255–278 (2009)
Byun, J. et al. Myocardial injury-induced fibroblast proliferation facilitates retroviral-mediated gene transfer to the rat heart in vivo . J. Gene Med. 2, 2–10 (2000)
Bhowmick, N. A. et al. TGF-β signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science 303, 848–851 (2004)
Zeisberg, E. M. et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nature Med. 13, 952–961 (2007)
Schneider, M. et al. S100A4 is upregulated in injured myocardium and promotes growth and survival of cardiac myocytes. Cardiovasc. Res. 75, 40–50 (2007)
Loffredo, F. S. et al. Bone marrow-derived cell therapy stimulates endogenous cardiomyocyte progenitors and promotes cardiac repair. Cell Stem Cell 8, 389–398 (2011)
Beyer, E. C. Gap junctions. Int. Rev. Cytol. 137C, 1–37 (1993)
Saffitz, J. E., Green, K. G., Kraft, W. J., Schechtman, K. B. & Yamada, K. A. Effects of diminished expression of connexin43 on gap junction number and size in ventricular myocardium. Am. J. Physiol. Heart Circ. Physiol. 278, H1662–H1670 (2000)
Acharya, A., Baek, S. T., Banfi, S., Eskiocak, B. & Tallquist, M. D. Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis 49, 870–877 (2011)
Hsieh, P. C. et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nature Med. 13, 970–974 (2007)
Zhou, Q., Brown, J., Kanarek, A., Rajagopal, J. & Melton, D. A. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455, 627–632 (2008)
Tandan, S. et al. Physical and functional interaction between calcineurin and the cardiac L-type Ca2+ channel. Circ. Res. 105, 51–60 (2009)
Laugwitz, K. L. et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 433, 647–653 (2005)
Luo, X. et al. Aberrant localization of intracellular organelles, Ca2+ signaling, and exocytosis in Mist1 null mice. J. Biol. Chem. 280, 12668–12675 (2005)
Grynkiewicz, G., Poenie, M. & Tsien, R. Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450 (1985)
Hardy, M. E. et al. Validation of a voltage-sensitive dye (di-4-ANEPPS)-based method for assessing drug-induced delayed repolarisation in beagle dog left ventricular midmyocardial myocytes. J. Pharmacol. Toxicol. Methods 60, 94–106 (2009)
Kitamura, T. et al. Efficient screening of retroviral cDNA expression libraries. Proc. Natl Acad. Sci. USA 92, 9146–9150 (1995)
Subramaniam, A. et al. Tissue-specific regulation of the α-myosin heavy chain gene promoter in transgenic mice. J. Biol. Chem. 266, 24613–24620 (1991)
Sohal, D. S. et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 89, 20–25 (2001)
Russell, J. L. et al. A dynamic notch injury response activates epicardium and contributes to fibrosis repair. Circ. Res. 108, 51–59 (2011)
Acknowledgements
We thank J. Cabrera for graphics. We are grateful to members of the Olson lab for critical reading of the manuscript. We thank D. Sosic, W. Tang, J. O’Rourke, N. Liu, M. Xin, A. Johnson and J. McAnally for discussions; J. Shelton and J. A. Richardson for histology. We are grateful to I. Bezprozvanny for the PTI Ca2+ Imaging System, and D. Srivastava for lentiviral plasmids. We thank the microarray core at University of Texas Southwestern Medical Center for collecting gene expression data. E.N.O. is supported by grants from NIH, the Donald W. Reynolds Center for Clinical Cardiovascular Research, the Robert A. Welch Foundation (grant I-0025), the Leducq Fondation-Transatlantic Network of Excellence in Cardiovascular Research Program, the American Heart Association-Jon Holden DeHaan Foundation and the Cancer Prevention & Research Institute of Texas (CPRIT).
Author information
Authors and Affiliations
Contributions
K.S. and E.N.O. conceived the project. K.S., Y.-J.N. and E.N.O. designed the experiments. K.S., Y.-J.N., X.L., X.Q., W.T., G.N.H., C.L.S. and A.A. performed experiments. J.A.H. contributed to surgical experiments; E.G.N. made the Fsp1-Cre mouse line. K.S. and R.B.-D. wrote the animal protocol. M.D.T. and R.B.-D. contributed scientific discussion. K.S., Y.-J.N., X.L., M.D.T., R.B.-D. and E.N.O. analysed data and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-21. (PDF 1479 kb)
Supplementary Movie 1
This movie shows beating induced cardiac-like myocytes (iCLM) generated from adult cardiac fibroblasts (CFs) 6 weeks post-infection of retroviruses expressing GHMT transcription factors. (MOV 2392 kb)
Supplementary Movie 2
This movie shows beating induced cardiac-like myocytes (iCLM) generated from adult cardiac fibroblasts (CFs) 6 weeks post-infection of retroviruses expressing GHMT transcription factors. (MOV 1796 kb)
Supplementary Movie 3
This movie shows beating induced cardiac-like myocytes (iCLM) generated from adult tail tip fibroblasts (TTFs) 6 weeks post-infection of retroviruses expressing GHMT transcription factors. (MOV 2004 kb)
Supplementary Movie 4
This movie shows beating induced cardiac-like myocytes (iCLM) that were isolated from mouse heart at 5 weeks post-MI and injection of retroviruses expressing GHMT transcription factors. (MOV 2628 kb)
Rights and permissions
About this article
Cite this article
Song, K., Nam, YJ., Luo, X. et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485, 599–604 (2012). https://doi.org/10.1038/nature11139
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11139
This article is cited by
-
Vitamin C facilitates direct cardiac reprogramming by inhibiting reactive oxygen species
Stem Cell Research & Therapy (2024)
-
Regeneration of the heart: from molecular mechanisms to clinical therapeutics
Military Medical Research (2023)
-
ETV2/ER71, the key factor leading the paths to vascular regeneration and angiogenic reprogramming
Stem Cell Research & Therapy (2023)
-
Translational landscape of direct cardiac reprogramming reveals a role of Ybx1 in repressing cardiac fate acquisition
Nature Cardiovascular Research (2023)
-
Secondary structures that regulate mRNA translation provide insights for ASO-mediated modulation of cardiac hypertrophy
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.