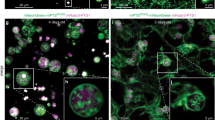
Abstract
Peroxisomes are eukaryotic organelles important for the metabolism of long-chain fatty acids1,2. Here we show that in numerous fungal species, several core enzymes of glycolysis, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and 3-phosphoglycerate kinase (PGK), reside in both the cytoplasm and peroxisomes. We detected in these enzymes cryptic type 1 peroxisomal targeting signals (PTS1)3, which are activated by post-transcriptional processes. Notably, the molecular mechanisms that generate the peroxisomal isoforms vary considerably among different species. In the basidiomycete plant pathogen Ustilago maydis, peroxisomal targeting of Pgk1 results from ribosomal read-through, whereas alternative splicing generates the PTS1 of Gapdh. In the filamentous ascomycete Aspergillus nidulans, peroxisomal targeting of these enzymes is achieved by exactly the opposite mechanisms. We also detected PTS1 motifs in the glycolytic enzymes triose-phosphate isomerase and fructose-bisphosphate aldolase. U. maydis mutants lacking the peroxisomal isoforms of Gapdh or Pgk1 showed reduced virulence. In addition, mutational analysis suggests that GAPDH, together with other peroxisomal NADH-dependent dehydrogenases, has a role in redox homeostasis. Owing to its hidden nature, partial peroxisomal targeting of well-studied cytoplasmic enzymes has remained undetected. Thus, we anticipate that further bona fide cytoplasmic proteins exhibit similar dual targeting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
De Duve, C. & Baudhuin, P. Peroxisomes (microbodies and related particles). Physiol. Rev. 46, 323–357 (1966)
Poirier, Y., Antonenkov, V. D., Glumoff, T. & Hiltunen, J. K. Peroxisomal β-oxidation – a metabolic pathway with multiple functions. Biochim. Biophys. Acta 1763, 1413–1426 (2006)
Gould, S. J., Keller, G. A., Hosken, N., Wilkinson, J. & Subramani, S. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108, 1657–1664 (1989)
Gabaldon, T. Peroxisome diversity and evolution. Phil. Trans. R. Soc. Lond. B 365, 765–773 (2010)
Graham, I. A. Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142 (2008)
Michels, P. A., Bringaud, F., Herman, M. & Hannaert, V. Metabolic functions of glycosomes in trypanosomatids. Biochim. Biophys. Acta 1763, 1463–1477 (2006)
Jedd, G. Fungal evo-devo: organelles and multicellular complexity. Trends Cell Biol. 21, 12–19 (2011)
Purdue, P. E. & Lazarow, P. B. Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17, 701–752 (2001)
Hoepfner, D., Schildknegt, D., Braakman, I., Philippsen, P. & Tabak, H. F. Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85–95 (2005)
Tabak, H. F. et al. Formation of peroxisomes: present and past. Biochim. Biophys. Acta 1763, 1647–1654 (2006)
Girzalsky, W., Saffian, D. & Erdmann, R. Peroxisomal protein translocation. Biochim. Biophys. Acta 1803, 724–731 (2010)
Brocard, C. & Hartig, A. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 (2006)
Lazarow, P. B. The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta 1763, 1599–1604 (2006)
Neuberger, G., Maurer-Stroh, S., Eisenhaber, B., Hartig, A. & Eisenhaber, F. Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 328, 581–592 (2003)
Jones, J. M., Nau, K., Geraghty, M. T., Erdmann, R. & Gould, S. J. Identification of peroxisomal acyl-CoA thioesterases in yeast and humans. J. Biol. Chem. 274, 9216–9223 (1999)
Baranov, P. V., Gesteland, R. F. & Atkins, J. F. Recoding: translational bifurcations in gene expression. Gene 286, 187–201 (2002)
Gruber, A. R., Lorenz, R., Bernhart, S. H., Neubock, R. & Hofacker, I. L. The Vienna RNA websuite. Nucleic Acids Res. 36, W70–W74 (2008)
Gurvitz, A. et al. The Saccharomyces cerevisiae peroxisomal 2,4-dienoyl-CoA reductase is encoded by the oleate-inducible gene SPS19 . J. Biol. Chem. 272, 22140–22147 (1997)
Managadze, D. et al. A proteomic approach towards the identification of the matrix protein content of the two types of microbodies in Neurospora crassa . Proteomics 10, 3222–3234 (2010)
Idnurm, A., Giles, S. S., Perfect, J. R. & Heitman, J. Peroxisome function regulates growth on glucose in the basidiomycete fungus Cryptococcus neoformans . Eukaryot. Cell 6, 60–72 (2007)
Kiel, J. A., Veenhuis, M. & van der Klei, I. J. PEX genes in fungal genomes: common, rare or redundant. Traffic 7, 1291–1303 (2006)
van Roermund, C. W., Elgersma, Y., Singh, N., Wanders, R. J. & Tabak, H. F. The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J. 14, 3480–3486 (1995)
Visser, W. F., van Roermund, C. W., Ijlst, L., Waterham, H. R. & Wanders, R. J. Metabolite transport across the peroxisomal membrane. Biochem. J. 401, 365–375 (2007)
Rottensteiner, H. & Theodoulou, F. L. The ins and outs of peroxisomes: co-ordination of membrane transport and peroxisomal metabolism. Biochim. Biophys. Acta 1763, 1527–1540 (2006)
Bernhardt, K., Wilkinson, S., Weber, A. P. & Linka, N. A peroxisomal carrier delivers NAD and contributes to optimal fatty acid degradation during storage oil mobilization. Plant J. 69, 1–13 (2011)
Kiel, J. A. et al. Matching the proteome to the genome: the microbody of penicillin-producing Penicillium chrysogenum cells. Funct. Integr. Genomics 9, 167–184 (2009)
Strijbis, K., van den Burg, J., F, Visser, W., van den Berg, M. & Distel, B. Alternative splicing directs dual localization of Candida albicans 6-phosphogluconate dehydrogenase to cytosol and peroxisomes. FEMS Yeast Res. 12, 61–68 (2012)
Kabran, P., Rossignol, T., Gaillardin, C., Nicaud, J. M. & Neuveglise, C. Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica . DNA Res. 10.1093/dnares/dss007 (24 February 2012)
Teusink, B., Walsh, M. C., van Dam, K. & Westerhoff, H. V. The danger of metabolic pathways with turbo design. Trends Biochem. Sci. 23, 162–169 (1998)
Haanstra, J. R. et al. Compartmentation prevents a lethal turbo-explosion of glycolysis in trypanosomes. Proc. Natl Acad. Sci. USA 105, 17718–17723 (2008)
Sambrook, J. & Russell, D. Molecular Cloning: a Laboratory Manual 3rd edn (Cold Spring Harbor Laboratory Press, 2001)
Schulz, B. et al. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60, 295–306 (1990)
Loubradou, G., Brachmann, A., Feldbrügge, M. & Kahmann, R. A homologue of the transcriptional repressor Ssn6p antagonizes cAMP signalling in Ustilago maydis . Mol. Microbiol. 40, 719–730 (2001)
Guthrie, C. & Fink, G. R. Guide to Yeast Genetics and Molecular and Cell Biology. (Academic, 2002)
Brachmann, A., König, J., Julius, C. & Feldbrügge, M. A reverse genetic approach for generating gene replacement mutants in Ustilago maydis . Mol. Genet. Genomics 272, 488 (2004)
Schink, K. O. & Bölker, M. Coordination of cytokinesis and cell separation by endosomal targeting of a Cdc42-specific guanine nucleotide exchange factor in Ustilago maydis . Mol. Biol. Cell 20, 1081–1088 (2009)
Spellig, T., Bottin, A. & Kahmann, R. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis . Mol. Gen. Genet. 252, 503–509 (1996)
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004)
Hoffman, C. S. & Winston, F. A ten minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation in Escherichia coli . Gene 57, 267–272 (1987)
Kämper, J. et al. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis . Nature 444, 97–101 (2006)
Acknowledgements
We are indebted to J. Heitman and W. K. Holloman for reading of the manuscript. We thank R. Fischer and G. Barth for providing strains and DNA. We acknowledge technical assistance by M. Piscator. We thank D. Lanver and T. Stehlik for discussions. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG-GK1216). J.A. received a fellowship from the Marburg University Research Academy (MARA).
Author information
Authors and Affiliations
Contributions
M.B. and J.F. designed this study. J.F. and J.A. performed the experiments. All authors contributed to data analysis. M.B. and J.F. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-18 and Supplementary Tables 1-2. (PDF 5328 kb)
Rights and permissions
About this article
Cite this article
Freitag, J., Ast, J. & Bölker, M. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 485, 522–525 (2012). https://doi.org/10.1038/nature11051
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11051
This article is cited by
-
Host-pathogen interaction between pitaya and Neoscytalidium dimidiatum reveals the mechanisms of immune response associated with defense regulators and metabolic pathways
BMC Plant Biology (2024)
-
Evasion of plant immunity by microbial pathogens
Nature Reviews Microbiology (2022)
-
Nonsense suppression therapies in human genetic diseases
Cellular and Molecular Life Sciences (2021)
-
Peroxisomal targeting of a protein phosphatase type 2C via mitochondrial transit
Nature Communications (2020)
-
The causes of evolvability and their evolution
Nature Reviews Genetics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.