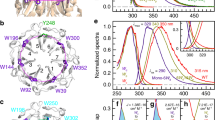
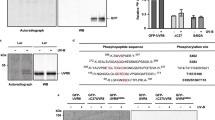
Abstract
The Arabidopsis thaliana protein UVR8 is a photoreceptor for ultraviolet-B. Upon ultraviolet-B irradiation, UVR8 undergoes an immediate switch from homodimer to monomer, which triggers a signalling pathway for ultraviolet protection. The mechanism by which UVR8 senses ultraviolet-B remains largely unknown. Here we report the crystal structure of UVR8 at 1.8 Å resolution, revealing a symmetric homodimer of seven-bladed β-propeller that is devoid of any external cofactor as the chromophore. Arginine residues that stabilize the homodimeric interface, principally Arg 286 and Arg 338, make elaborate intramolecular cation–π interactions with surrounding tryptophan amino acids. Two of these tryptophans, Trp 285 and Trp 233, collectively serve as the ultraviolet-B chromophore. Our structural and biochemical analyses identify the molecular mechanism for UVR8-mediated ultraviolet-B perception, in which ultraviolet-B radiation results in destabilization of the intramolecular cation–π interactions, causing disruption of the critical intermolecular hydrogen bonds mediated by Arg 286 and Arg 338 and subsequent dissociation of the UVR8 homodimer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Falciatore, A. & Bowler, C. The evolution and function of blue and red light photoreceptors. Curr. Top. Dev. Biol. 68, 317–350 (2005)
van der Linden, A. M. et al. Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 8, e1000503 (2010)
Fogle, K. J., Parson, K. G., Dahm, N. A. & Holmes, T. C. CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science 331, 1409–1413 (2011)
Jiao, Y., Lau, O. S. & Deng, X. W. Light-regulated transcriptional networks in higher plants. Nature Rev. Genet. 8, 217–230 (2007)
Kami, C., Lorrain, S., Hornitschek, P. & Fankhauser, C. Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91, 29–66 (2010)
Quail, P. H. Phytochrome photosensory signalling networks. Nature Rev. Mol. Cell Biol. 3, 85–93 (2002)
Briggs, W. R. & Christie, J. M. Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 7, 204–210 (2002)
Chaves, I. et al. The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62, 335–364 (2011)
Cashmore, A. R., Jarillo, J. A., Wu, Y. J. & Liu, D. Cryptochromes: blue light receptors for plants and animals. Science 284, 760–765 (1999)
Christie, J. M. Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58, 21–45 (2007)
Liu, H., Liu, B., Zhao, C., Pepper, M. & Lin, C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 16, 684–691 (2011)
Rizzini, L. et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106 (2011)
Möglich, A., Yang, X., Ayers, R. A. & Moffat, K. Structure and function of plant photoreceptors. Annu. Rev. Plant Biol. 61, 21–47 (2010)
Kliebenstein, D. J., Lim, J. E., Landry, L. G. & Last, R. L. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130, 234–243 (2002)
Brown, B. A. et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl Acad. Sci. USA 102, 18225–18230 (2005)
Brown, B. A. & Jenkins, G. I. UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 146, 576–588 (2008)
Kaiserli, E. & Jenkins, G. I. UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19, 2662–2673 (2007)
Li, D. & Roberts, R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 58, 2085–2097 (2001)
Wall, M. A. et al. The structure of the G protein heterotrimer Giα1β1γ2 . Cell 83, 1047–1058 (1995)
Renault, L. et al. The 1.7 Å crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 392, 97–101 (1998)
Gallivan, J. P. & Dougherty, D. A. Cation–π interactions in structural biology. Proc. Natl Acad. Sci. USA 96, 9459–9464 (1999)
Gallivan, J. P. & Dougherty, D. A. A computational study of cation−π interactions vs salt bridges in aqueous media: implications for protein engineering. J. Am. Chem. Soc. 122, 870–874 (2000)
Dougherty, D. A. Cation−π interactions involving aromatic amino acids. J. Nutr. 137, 1504S–1508S (2007)
Sinnokrot, M. O., Valeev, E. F. & Sherrill, C. D. Estimates of the ab initio limit for π–π interactions: the benzene dimer. J. Am. Chem. Soc. 124, 10887–10893 (2002)
Wagner, J. R., Brunzelle, J. S., Forest, K. T. & Vierstra, R. D. A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 (2005)
Ulijasz, A. T. et al. Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature 463, 250–254 (2010)
Brudler, R. et al. Identification of a new cryptochrome class. Structure, function, and evolution. Mol. Cell 11, 59–67 (2003)
Brautigam, C. A. et al. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 101, 12142–12147 (2004)
Crosson, S. & Moffat, K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl Acad. Sci. USA 98, 2995–3000 (2001)
Crosson, S. & Moffat, K. Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell 14, 1067–1075 (2002)
Yu, H. T., Colucci, W. J., McLaughlin, M. L. & Barkley, M. D. Fluorescence quenching in indoles by excited-state proton transfer. J. Am. Chem. Soc. 114, 8449–8454 (1992)
Christie, J. M. et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Sciencehttp://dx.doi.org/10.1126/science.1218091 (9 February 2012)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 (2002)
Cowtan, K. The Buccaneer software for automated model building. Acta Crystallogr. D 62, 1002–1011 (2006)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
DeLano, W. L. The PyMOL Molecular Graphics System. http://www.pymol.org (2002)
Acknowledgements
We thank J. He and S. Huang at SSRF, and K. Hasegawa and T. Kumasaka at the SPring-8 beamline BL41XU, for assistance. This work was supported by funds from the Ministry of Science and Technology (grant no. 2009CB918801 to Y.S., and 2012CB910900 to X.W.D.), the National Natural Science Foundation, and the Beijing Municipal Commissions of Education and Science and Technology.
Author information
Authors and Affiliations
Contributions
D.W., Q.H., H.D., X.W.D. and Y.S. designed all experiments. D.W., Q.H., Z.Y., W.C., C.Y. and J.Z. performed the experiments. D.W., Q.H., Z.Y., W.C., C.Y., X.H., J.Z., P.Y., H.D., J.W., X.W.D. and Y.S. contributed to technical work and data analysis. D.W., Q.H., Z.Y., W.C., C.Y., J.W., X.W.D. and Y.S. contributed to manuscript preparation. Y.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-14 with legends, Supplementary Table 1 and additional references. (PDF 1491 kb)
Rights and permissions
About this article
Cite this article
Di Wu, Hu, Q., Yan, Z. et al. Structural basis of ultraviolet-B perception by UVR8. Nature 484, 214–219 (2012). https://doi.org/10.1038/nature10931
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10931
This article is cited by
-
How is UVR8 relevant in plants? New evidence
Plant Growth Regulation (2023)
-
On the concepts and correct use of radiometric quantities for assessing the light environment and their application to plant research
Biophysical Reviews (2023)
-
Einfluss der Verschmutzung von Reflexionsfolien auf ihr Reflexionsverhalten
Erwerbs-Obstbau (2023)
-
Dynamics and mechanism of dimer dissociation of photoreceptor UVR8
Nature Communications (2022)
-
Light-Quality Manipulation to Control Plant Growth and Photomorphogenesis in Greenhouse Horticulture: The State of the Art and the Opportunities of Modern LED Systems
Journal of Plant Growth Regulation (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.