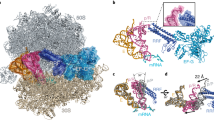
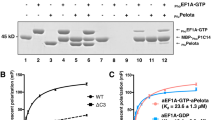
Abstract
Ribosome-driven protein biosynthesis is comprised of four phases: initiation, elongation, termination and recycling. In bacteria, ribosome recycling requires ribosome recycling factor and elongation factor G, and several structures of bacterial recycling complexes have been determined. In the eukaryotic and archaeal kingdoms, however, recycling involves the ABC-type ATPase ABCE1 and little is known about its structural basis. Here we present cryo-electron microscopy reconstructions of eukaryotic and archaeal ribosome recycling complexes containing ABCE1 and the termination factor paralogue Pelota. These structures reveal the overall binding mode of ABCE1 to be similar to canonical translation factors. Moreover, the iron–sulphur cluster domain of ABCE1 interacts with and stabilizes Pelota in a conformation that reaches towards the peptidyl transferase centre, thus explaining how ABCE1 may stimulate peptide-release activity of canonical termination factors. Using the mechanochemical properties of ABCE1, a conserved mechanism in archaea and eukaryotes is suggested that couples translation termination to recycling, and eventually to re-initiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Winzeler, E. A. et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999)
Coelho, C. M. et al. Growth and cell survival are unevenly impaired in pixie mutant wing discs. Development 132, 5411–5424 (2005)
Estevez, A. M., Haile, S., Steinbuchel, M., Quijada, L. & Clayton, C. Effects of depletion and overexpression of the Trypanosoma brucei ribonuclease L inhibitor homologue. Mol. Biochem. Parasitol. 133, 137–141 (2004)
Pisarev, A. V. et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell 37, 196–210 (2010)
Pisareva, V. P., Skabkin, M. A., Hellen, C. U., Pestova, T. V. & Pisarev, A. V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 30, 1804–1817 (2011)
Karcher, A., Schele, A. & Hopfner, K.-P. X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J. Biol. Chem. 283, 7962–7971 (2008)
Karcher, A., Buttner, K., Martens, B., Jansen, R. P. & Hopfner, K. P. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure 13, 649–659 (2005)
Barthelme, D. et al. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc. Natl. Acad. Sci. USA 108, 3228–3233 (2011)
Barthelme, D. et al. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J. Biol. Chem. 282, 14598–14607 (2007)
Dong, J. et al. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem. 279, 42157–42168 (2004)
Andersen, D. & Leevers, S. The essential Drosophila ATP-binding cassette domain protein, Pixie, binds the 40S ribosome in an ATP-dependent manner and is required for translation initiation. J. Biol. Chem. 282, 14752–14760 (2007)
Shoemaker, C. J. & Green, R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl Acad. Sci. USA 108, E1392–E1398 (2011)
Khoshnevis, S. et al. The iron–sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 11, 214–219 (2010)
Doma, M. & Parker, R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564 (2006)
Atkinson, G., Baldauf, S. & Hauryliuk, V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol. Biol. 8, 290 (2008)
Lee, H. H. et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol. Cell 27, 938–950 (2007)
Frischmeyer, P. et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–2261 (2002)
van Hoof, A., Frischmeyer, P. A., Dietz, H. C. & Parker, R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264 (2002)
Becker, T. et al. Structure of the no-go mRNA decay complex Dom34–Hbs1 bound to a stalled 80S ribosome. Nature Struct. Mol. Biol. 18, 715–720 (2011)
Shoemaker, C. J., Eyler, D. E. & Green, R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330, 369–372 (2010)
Hosoda, N. et al. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem. 278, 38287–38291 (2003)
Endoh, T., Kanai, T. & Imanaka, T. A highly productive system for cell-free protein synthesis using a lysate of the hyperthermophilic archaeon, Thermococcus kodakaraensis. Appl. Microbiol. Biotechnol. 74, 1153–1161 (2007)
Rabl, J., Leibundgut, M., Ataide, S. F., Haag, A. & Ban, N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331, 730–736 (2011)
Armache, J. P. et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. Proc. Natl Acad. Sci. USA 107, 19754–19759 (2010)
Armache, J. P. et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proc. Natl Acad. Sci. USA 107, 19748–19753 (2010)
Schmeing, T. M. et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 (2009)
Gao, Y. G. et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699 (2009)
Taylor, D. J. et al. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 26, 2421–2431 (2007)
Spahn, C. M. et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 23, 1008–1019 (2004)
Villa, E. et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl Acad. Sci. USA 106, 1063–1068 (2009)
Connell, S. R. et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol. Cell 25, 751–764 (2007)
Kobayashi, K. et al. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1α complex. Proc. Natl Acad. Sci. USA 107, 17575–17579 (2010)
Frolova, L. et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5, 1014–1020 (1999)
Song, H. et al. The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100, 311–321 (2000)
Hopfner, K. P. & Tainer, J. A. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 13, 249–255 (2003)
Smith, P. C. et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 (2002)
Locher, K. P. Structure and mechanism of ATP-binding cassette transporters. Phil. Trans. R. Soc. Lond. B 364, 239–245 (2009)
Pisarev, A. V., Hellen, C. U. & Pestova, T. V. Recycling of eukaryotic posttermination ribosomal complexes. Cell 131, 286–299 (2007)
Kispal, G. et al. Biogenesis of cytosolic ribosomes requires the essential iron–sulphur protein Rli1p and mitochondria. EMBO J. 24, 589–598 (2005)
Yarunin, A. et al. Functional link between ribosome formation and biogenesis of iron–sulfur proteins. EMBO J. 24, 580–588 (2005)
Pavlov, M. Y., Antoun, A., Lovmar, M. & Ehrenberg, M. Complementary roles of initiation factor 1 and ribosome recycling factor in 70S ribosome splitting. EMBO J. 27, 1706–1717 (2008)
Beckmann, R. et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 (2001)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Soding, J., Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005)
Eswar, N., Eramian, D., Webb, B., Shen, M. Y. & Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 (2008)
Endoh, T. et al. Cell-free protein synthesis at high temperatures using the lysate of a hyperthermophile. J. Biotechnol. 126, 186–195 (2006)
Endoh, T., Kanai, T. & Imanaka, T. Effective approaches for the production of heterologous proteins using the Thermococcus kodakaraensis-based translation system. J. Biotechnol. 133, 177–182 (2008)
Graille, M., Chaillet, M. & van Tilbeurgh, H. Structure of yeast Dom34: a protein related to translation termination factor eRF1 and involved in no-go decay. J. Biol. Chem. 283, 7145–7154 (2008)
Lee, H. H. et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol. Cell 27, 938–950 (2007)
Chen, J. Z. & Grigorieff, N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J. Struct. Biol. 157, 168–173 (2007)
Becker, T. et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326, 1369–1373 (2009)
Chen, L. et al. Structure of the Dom34–Hbs1 complex and implications for no-go decay. Nature Struct. Mol. Biol. 17, 1233–1240 (2010)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Pettersen, E. F. et al. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Schröder, G. F., Brunger, A. T. & Levitt, M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure 15, 1630–1641 (2007)
Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008)
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005)
Acknowledgements
We thank A. Schele and A. Gilmozzi for technical assistance, and D. Wilson for critical discussions. This work was supported by grants from the Deutsche Forschungsgemeinschaft, SFB594 (to R.B.), SFB646 (to T.B., R.B. and K.-P.H.), National Institutes of Health U19 AI083025 (to K.-P.H.) and by the Fonds der chemischen Industrie (to S.F.).
Author information
Authors and Affiliations
Contributions
T.B. and R.B. designed the study, T.B. processed the yeast SL–RNC–Dom34–Rli1 complex and interpreted the cryo-EM structures, S.F. purified archaeal proteins and reconstituted the archaeal 70S–aPelota–aABCE1 sample, processed all archaeal data sets and interpreted the cryo-EM structures, S.W. developed an automated workflow for data processing from the Titan Krios microscope, C.J.S. purified yeast Rli1p, A.M.A. built the archaeal 70S ribosome rRNA model, J.-P.A. built the archaeal 70S ribosome protein models, H.S. reconstituted the SL–RNC–Dom34–Rli1 sample, C.U. prepared cryo-EM grids and assisted in data collection, O.B. optimized and performed cryo-EM data collection, I.D. implemented software for automated data collection on the Titan Krios microscope, A.K. purified archaeal ABCE1, M.T. provided P. furiosus and Thermococcus kodakaraensis cells, T.B., S.F., K.-P.H., R.G. and R.B. interpreted results.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-5 with legends, Supplementary Tables 1-2, legends for Supplementary Movies 1-3 and additional references. (PDF 1939 kb)
Supplementary Movie 1
This movie shows the domain movement of Pelota central domain stabilized by ABCE1 – yeast (see Supplementary Information file for full legend). (MOV 14182 kb)
Supplementary Movie 2
This movie shows the domain movement of Pelota central domain stabilized by ABCE1 - archaea (see Supplementary Information file for full legend). (MOV 10126 kb)
Supplementary Movie 3
This move shows the conformational transition of ABCE1 (see Supplementary Information file for full legend). (MOV 14925 kb)
Rights and permissions
About this article
Cite this article
Becker, T., Franckenberg, S., Wickles, S. et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482, 501–506 (2012). https://doi.org/10.1038/nature10829
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10829
This article is cited by
-
Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2
Nature Communications (2023)
-
DNA-free genome editing in tomato protoplasts using CRISPR/Cas9 ribonucleoprotein delivery
Horticulture, Environment, and Biotechnology (2023)
-
An overview of 25 years of research on Thermococcus kodakarensis, a genetically versatile model organism for archaeal research
Folia Microbiologica (2020)
-
Pelota-interacting G protein Hbs1 is required for spermatogenesis in Drosophila
Scientific Reports (2019)
-
Application of Whole Genome Resequencing in Mapping of a Tomato Yellow Leaf Curl Virus Resistance Gene
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.