Abstract
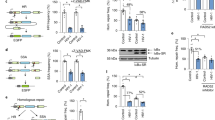
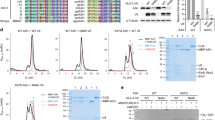
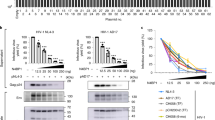
The human APOBEC3 cytidine deaminases are potent inhibitors of diverse retroviruses, including human immunodeficiency virus-1 (HIV-1)1,2,3,4,5,6. HIV-1 Vif forms an E3 ubiquitin ligase complex with cullin 5 (CUL5), elongin B and elongin C 7,8,9, which promotes the polyubiquitination and degradation of APOBEC3 substrates7,10,11,12,13,14. Here we demonstrate in human T cells that core binding factor β (CBF-β) is a key regulator of the evasion of HIV-1 from the host defence mediated by APOBEC3. CBF-β, the non-DNA-binding subunit of a heterodimeric transcription factor, regulates the folding and DNA-binding activity of partner RUNX family proteins, which have important roles in the development and differentiation of diverse cell types, including T lymphocytes15,16. In our study, knockdown of endogenous CBF-β blocked Vif-induced APOBEC3G polyubiquitination and degradation. CBF-β was not required for the interaction between Vif and APOBEC3G, yet was essential for the assembly of the Vif–CUL5 E3-ubiquitin-ligase complex. CBF-β proved to be a unique regulator of primate lentiviral Vif and not a general component of the CUL5 E3 ubiquitin ligase. We show that Vif and CBF-β physically interact, and that the amino-terminal region of Vif is required for this interaction. Furthermore, interactions with Vif required regions in CBF-β that are not involved in RUNX protein binding17,18,19. Considering the importance of the interaction between Vif and CBF-β, disrupting this interaction represents an attractive pharmacological intervention against HIV-1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002)
Navarro, F. & Landau, N. R. Recent insights into HIV-1 Vif. Curr. Opin. Immunol. 16, 477–482 (2004)
Cullen, B. R. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80, 1067–1076 (2006)
Chiu, Y. L. & Greene, W. C. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26, 317–353 (2008)
Malim, M. H. & Emerman, M. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3, 388–398 (2008)
Goila-Gaur, R. & Strebel, K. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5, 51 (2008)
Yu, X. et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-CUL5-SCF complex. Science 302, 1056–1060 (2003)
Mehle, A., Goncalves, J., Santa-Marta, M., McPike, M. & Gabuzda, D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-CUL5 complex that promotes APOBEC3G degradation. Genes Dev. 18, 2861–2866 (2004)
Yu, Y., Xiao, Z., Ehrlich, E. S., Yu, X. & Yu, X. F. Selective assembly of HIV-1 Vif-CUL5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18, 2867–2872 (2004)
Stopak, K., de Noronha, C., Yonemoto, W. & Greene, W. C. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12, 591–601 (2003)
Marin, M., Rose, K. M., Kozak, S. L. & Kabat, D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nature Med. 9, 1398–1403 (2003)
Conticello, S. G., Harris, R. S. & Neuberger, M. S. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13, 2009–2013 (2003)
Sheehy, A. M., Gaddis, N. C. & Malim, M. H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nature Med. 9, 1404–1407 (2003)
Mehle, A. et al. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279, 7792–7798 (2003)
de Bruijn, M. F. & Speck, N. A. Core-binding factors in hematopoiesis and immune function. Oncogene 23, 4238–4248 (2004)
Ito, Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv. Cancer Res. 99, 33–76 (2008)
Tahirov, T. H. et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFβ. Cell 104, 755–767 (2001)
Bravo, J., Li, Z., Speck, N. A. & Warren, A. J. The leukemia-associated AML1 (Runx1)–CBFβ complex functions as a DNA-induced molecular clamp. Nature Struct. Biol. 8, 371–378 (2001)
Yan, J., Liu, Y., Lukasik, S. M., Speck, N. A. & Bushweller, J. H. CBFβ allosterically regulates the Runx1 Runt domain via a dynamic conformational equilibrium. Nature Struct. Mol. Biol. 11, 901–906 (2004)
Liu, B., Sarkis, P. T., Luo, K., Yu, Y. & Yu, X. F. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-CUL5-ElonB/C E3 ubiquitin ligase. J. Virol. 79, 9579–9587 (2005)
Simon, V. et al. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1, e6 (2005)
Tian, C. et al. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 80, 3112–3115 (2006)
Schröfelbauer, B., Senger, T., Manning, G. & Landau, N. R. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 80, 5984–5991 (2006)
Russell, R. A. & Pathak, V. K. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81, 8201–8210 (2007)
Mehle, A. et al. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J. Virol. 81, 13235–13241 (2007)
He, Z., Zhang, W., Chen, G., Xu, R. & Yu, X. F. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381, 1000–1011 (2008)
Zhang, W., Chen, G., Niewiadomska, A. M., Xu, R. & Yu, X. F. Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host anti-viral proteins. PLoS ONE 3, e3963 (2008)
Cao, W., Adya, N., Britos-Bray, M., Liu, P. P. & Friedman, A. D. The core binding factor (CBF) α interaction domain and the smooth muscle myosin heavy chain (SMMHC) segment of CBFβ-SMMHC are both required to slow cell proliferation. J. Biol. Chem. 273, 31534–31540 (1998)
Luo, K. et al. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5–E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl Acad. Sci. USA 102, 11444–11449 (2005)
Luo, K. et al. Adenovirus E4orf6 assembles with Cullin5-ElonginB-ElonginC E3 ubiquitin ligase through an HIV/SIV Vif-like BC-box to regulate p53. FASEB J. 21, 1742–1750 (2007)
Acknowledgements
We thank K. Strebel, A. Friedman, N. Speck, D. Yue, R. Siliciano, M. Malim, R. Harris, T. Inoue and D. Gabuzda for critical reagents; T. Wang, K. Zhao, X. Zhou and A. Zhen for technical assistance; R. Markham, J. Margolick and J. Bream for thoughtful discussions; and D. McClellan for editorial assistance. We also wish to thank staff within the Mass Spectrometry Core and the Institute for Basic Biomedical Sciences Microscope Facility at Johns Hopkins School of Medicine for their technical assistance. The following reagents were obtained through the AIDS Research and Reference Reagents Program, Division of AIDS, NIAID, NIH: monoclonal antibodies against HIV-1 p24 (B. Chesebro and H. Chen), pNL4-3 (M. Martin), pcDNA-hVif (S. Bour and K. Strebel), antiserum to HIV-1 Vif (D. Gabuzda) and MAGI-CCR5 cells (J. Overbaugh). This work was supported in part by funding from the Chinese Ministry of Science and Technology (2012CB911100) and Chinese Ministry of Education (IRT1016), the Key Laboratory of Molecular Virology, Jilin Province (20102209), China, and a grant (2R56AI62644-6) from the NIAID.
Author information
Authors and Affiliations
Contributions
W.Z., J.D., S.L.E. and Y.Y. performed experiments and analysed the data. X.-F.Y. directed the project, analysed the data, and wrote the paper with help from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
The file contains Supplementary Figures 1-9 with legends. (PDF 6268 kb)
Rights and permissions
About this article
Cite this article
Zhang, W., Du, J., Evans, S. et al. T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481, 376–379 (2012). https://doi.org/10.1038/nature10718
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10718
This article is cited by
-
Structural basis of HIV-1 Vif-mediated E3 ligase targeting of host APOBEC3H
Nature Communications (2023)
-
The E3 ligase RNF5 restricts SARS-CoV-2 replication by targeting its envelope protein for degradation
Signal Transduction and Targeted Therapy (2023)
-
RNA sensor MDA5 suppresses LINE-1 retrotransposition by regulating the promoter activity of LINE-1 5′-UTR
Mobile DNA (2022)
-
CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation
Nature Chemical Biology (2021)
-
Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G
Nature Communications (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.