Abstract
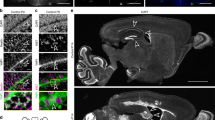
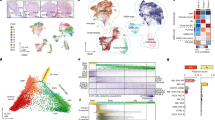
Understanding the biology that underlies histologically similar but molecularly distinct subgroups of cancer has proven difficult because their defining genetic alterations are often numerous, and the cellular origins of most cancers remain unknown1,2,3. We sought to decipher this heterogeneity by integrating matched genetic alterations and candidate cells of origin to generate accurate disease models. First, we identified subgroups of human ependymoma, a form of neural tumour that arises throughout the central nervous system (CNS). Subgroup-specific alterations included amplifications and homozygous deletions of genes not yet implicated in ependymoma. To select cellular compartments most likely to give rise to subgroups of ependymoma, we matched the transcriptomes of human tumours to those of mouse neural stem cells (NSCs), isolated from different regions of the CNS at different developmental stages, with an intact or deleted Ink4a/Arf locus (that encodes Cdkn2a and b). The transcriptome of human supratentorial ependymomas with amplified EPHB2 and deleted INK4A/ARF matched only that of embryonic cerebral Ink4a/Arf−/− NSCs. Notably, activation of Ephb2 signalling in these, but not other, NSCs generated the first mouse model of ependymoma, which is highly penetrant and accurately models the histology and transcriptome of one subgroup of human supratentorial tumour. Further, comparative analysis of matched mouse and human tumours revealed selective deregulation in the expression and copy number of genes that control synaptogenesis, pinpointing disruption of this pathway as a critical event in the production of this ependymoma subgroup. Our data demonstrate the power of cross-species genomics to meticulously match subgroup-specific driver mutations with cellular compartments to model and interrogate cancer subgroups.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Parsons, D. W. et al. An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 (2008)
Stommel, J. M. et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318, 287–290 (2007)
The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008)
Kleihues, P. et al. The WHO classification of tumors of the nervous system. J. Neuropathol. Exp. Neurol. 61, 215–225; discussion 226–219 (2002)
Merchant, T. E. et al. Conformal radiotherapy after surgery for paediatric ependymoma: a prospective study. Lancet Oncol. 10, 258–266 (2009)
Modena, P. et al. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J. Clin. Oncol. 24, 5223–5233 (2006)
Taylor, M. D. et al. Radial glia cells are candidate stem cells of ependymoma. Cancer Cell 8, 323–335 (2005)
Puget, S. et al. Candidate genes on chromosome 9q33–34 involved in the progression of childhood ependymomas. J. Clin. Oncol. 27, 1884–1892 (2009)
Diskin, S. J. et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature 459, 987–991 (2009)
Dejosez, M. et al. Ronin is essential for embryogenesis and the pluripotency of mouse embryonic stem cells. Cell 133, 1162–1174 (2008)
Nakano, I. et al. Phosphoserine phosphatase is expressed in the neural stem cell niche and regulates neural stem and progenitor cell proliferation. Stem Cells 25, 1975–1984 (2007)
Kayser, M. S., Nolt, M. J. & Dalva, M. B. EphB receptors couple dendritic filopodia motility to synapse formation. Neuron 59, 56–69 (2008)
Chess, A. Monoallelic expression of protocadherin genes. Nature Genet. 37, 120–121 (2005)
Cueni, L. et al. T-type Ca2+ channels, SK2 channels and SERCAs gate sleep-related oscillations in thalamic dendrites. Nature Neurosci. 11, 683–692 (2008)
Gogolla, N. et al. Wnt signaling mediates experience-related regulation of synapse numbers and mossy fiber connectivities in the adult hippocampus. Neuron 62, 510–525 (2009)
Geppert, M. et al. The small GTP-binding protein Rab3A regulates a late step in synaptic vesicle fusion. Nature 387, 810–814 (1997)
Taraska, J. W. & Almers, W. Bilayers merge even when exocytosis is transient. Proc. Natl Acad. Sci. USA 101, 8780–8785 (2004)
Dahlhaus, M. et al. Notch1 signaling in pyramidal neurons regulates synaptic connectivity and experience-dependent modifications of acuity in the visual cortex. J. Neurosci. 28, 10794–10802 (2008)
Kriegstein, A. & Alvarez-Buylla, A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184 (2009)
Merkle, F. T. et al. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc. Natl Acad. Sci. USA 101, 17528–17532 (2004)
Kriegstein, A. R. & Gotz, M. Radial glia diversity: a matter of cell fate. Glia 43, 37–43 (2003)
Anthony, T. E. et al. Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev. 19, 1028–1033 (2005)
Gong, S. et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 (2003)
Schmid, R. S., Yokota, Y. & Anton, E. S. Generation and characterization of brain lipid-binding protein promoter-based transgenic mouse models for the study of radial glia. Glia 53, 345–351 (2006)
Molofsky, A. V. et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature 443, 448–452 (2006)
McGuire, C. S., Sainani, K. L. & Fisher, P. G. Incidence patterns for ependymoma: a surveillance, epidemiology, and end results study. J. Neurosurg. 110, 725–729 (2009)
Genander, M. et al. Dissociation of EphB2 signaling pathways mediating progenitor cell proliferation and tumor suppression. Cell 139, 679–692 (2009)
Rousseau, E. et al. Trisomy 19 ependymoma, a newly recognized genetico-histological association, including clear cell ependymoma. Mol. Cancer 6, 47 (2007)
Phillips, H. S. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9, 157–173 (2006)
Thompson, M. C. et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J. Clin. Oncol. 24, 1924–1931 (2006)
Acknowledgements
R.J.G. holds the Howard C. Schott Research Chair from the Malia’s Cord Foundation, and is supported by grants from the National Institutes of Health (R01CA129541, P01CA96832 and P30CA021765), the Collaborative Ependymoma Research Network (CERN) and by the American Lebanese Syrian Associated Charities (ALSAC). K.D.W. is supported by NRSA Training Grant T32 CA070089. We are grateful to N. Heintz for providing the Blbp–eGFP mouse, T. Pawson for providing mouse Ephb2 cDNA, M. Roussel for the pCX4-IRES-RFP virus, J. Downing and M. Relling for access to 500K SNP profiles of normal human DNA, and the staff of the Hartwell center for Bioinformatics and Biotechnology, Vector Production Core, ARC, AIC and Cell and Tissue Imaging at St Jude Children’s Research Hospital for technical assistance.
Author information
Authors and Affiliations
Contributions
R.J.G., R.G.G. and M.D.T. conceived the research and planned experiments. R.A.J. and K.D.W. also planned and conducted the great majority of the experiments under the direction of R.J.G.; A.G., Y.G., J.C.A., M.D.T. and R.G.G. provided human tumour material. All authors contributed experimental expertise and participated in the writing of the manuscript. D.F., S.B.P., Y.-D.W. and G.N. provided bioinformatic expertise. D.W.E. provided pathology review and iFISH analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, References, Supplementary Figures 1-6 with legends, Supplementary Table legends 1-6 (see pages 32-33) and Supplementary Tables 1-6. (PDF 9737 kb)
Supplementary Information
This file contains gene cards, which contain a comprehensive summary of the DNA alteration and gene expression within each validated focal amplification and deletion identified in Figure 1 of the main paper. (PDF 24320 kb)
Rights and permissions
About this article
Cite this article
Johnson, R., Wright, K., Poppleton, H. et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature 466, 632–636 (2010). https://doi.org/10.1038/nature09173
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09173
This article is cited by
-
Targeting the key players of phenotypic plasticity in cancer cells by phytochemicals
Cancer and Metastasis Reviews (2024)
-
Nuclear condensates of YAP fusion proteins alter transcription to drive ependymoma tumourigenesis
Nature Cell Biology (2023)
-
Superenhancers as master gene regulators and novel therapeutic targets in brain tumors
Experimental & Molecular Medicine (2023)
-
Review and update on pediatric ependymoma
Child's Nervous System (2023)
-
Cell-lineage controlled epigenetic regulation in glioblastoma stem cells determines functionally distinct subgroups and predicts patient survival
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.