Abstract
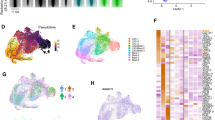
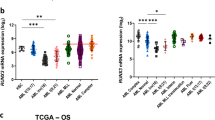
Chronic myelogenous leukaemia (CML) can progress from a slow growing chronic phase to an aggressive blast crisis phase1, but the molecular basis of this transition remains poorly understood. Here we have used mouse models of CML2,3 to show that disease progression is regulated by the Musashi–Numb signalling axis4,5. Specifically, we find that the chronic phase is marked by high levels of Numb expression whereas the blast crisis phase has low levels of Numb expression, and that ectopic expression of Numb promotes differentiation and impairs advanced-phase disease in vivo. As a possible explanation for the decreased levels of Numb in the blast crisis phase, we show that NUP98–HOXA9, an oncogene associated with blast crisis CML6,7, can trigger expression of the RNA-binding protein Musashi2 (Msi2), which in turn represses Numb. Notably, loss of Msi2 restores Numb expression and significantly impairs the development and propagation of blast crisis CML in vitro and in vivo. Finally we show that Msi2 expression is not only highly upregulated during human CML progression but is also an early indicator of poorer prognosis. These data show that the Musashi–Numb pathway can control the differentiation of CML cells, and raise the possibility that targeting this pathway may provide a new strategy for the therapy of aggressive leukaemias.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Calabretta, B. & Perrotti, D. The biology of CML blast crisis. Blood 103, 4010–4022 (2004)
Daley, G. Q., Van Etten, R. A. & Baltimore, D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science 247, 824–830 (1990)
Pear, W. S. et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood 92, 3780–3792 (1998)
Uemura, T., Shepherd, S., Ackerman, L., Jan, L. Y. & Jan, Y. N. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58, 349–360 (1989)
Nakamura, M., Okano, H., Blendy, J. A. & Montell, C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron 13, 67–81 (1994)
Mayotte, N., Roy, D. C., Yao, J., Kroon, E. & Sauvageau, G. Oncogenic interaction between BCR-ABL and NUP98-HOXA9 demonstrated by the use of an in vitro purging culture system. Blood 100, 4177–4184 (2002)
Dash, A. B. et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc. Natl Acad. Sci. USA 99, 7622–7627 (2002)
Witte, O. The role of Bcr-Abl in chronic myeloid leukemia and stem cell biology. Semin. Hematol. 38, 3–8 (2001)
Ren, R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nature Rev. Cancer 5, 172–183 (2005)
Melo, J. V. & Barnes, D. J. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nature Rev. Cancer 7, 441–453 (2007)
Goldman, J. M. & Melo, J. V. BCR-ABL in chronic myelogenous leukemia–how does it work? Acta Haematol. 119, 212–217 (2008)
Knoblich, J. A. Mechanisms of asymmetric cell division during animal development. Curr. Opin. Cell Biol. 9, 833–841 (1997)
Spana, E. P. & Doe, C. Q. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17, 21–26 (1996)
Shen, Q., Zhong, W., Jan, Y. N. & Temple, S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development 129, 4843–4853 (2002)
Wu, M. et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell 1, 541–554 (2007)
Wang, H., Ouyang, Y., Somers, W. G., Chia, W. & Lu, B. Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon. Nature 449, 96–100 (2007)
Neering, S. J. et al. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood 110, 2578–2585 (2007)
Justice, N., Roegiers, F., Jan, L. Y. & Jan, Y. N. Lethal giant larvae acts together with numb in notch inhibition and cell fate specification in the Drosophila adult sensory organ precursor lineage. Curr. Biol. 13, 778–783 (2003)
Wakamatsu, Y., Maynard, T. M., Jones, S. U. & Weston, J. A. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron 23, 71–81 (1999)
Colaluca, I. N. et al. NUMB controls p53 tumour suppressor activity. Nature 451, 76–80 (2008)
Imai, T. et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol. Cell. Biol. 21, 3888–3900 (2001)
Okabe, M., Imai, T., Kurusu, M., Hiromi, Y. & Okano, H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature 411, 94–98 (2001)
Sakakibara, S. et al. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc. Natl Acad. Sci. USA 99, 15194–15199 (2002)
Okano, H. et al. Function of RNA-binding protein Musashi-1 in stem cells. Exp. Cell Res. 306, 349–356 (2005)
Taniwaki, T. et al. Characterization of an exchangeable gene trap using pU-17 carrying a stop codon-βgeo cassette. Dev. Growth Differ. 47, 163–172 (2005)
Radich, J. P. et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc. Natl Acad. Sci. USA 103, 2794–2799 (2006)
Nakahara, F. et al. Hes1 immortalizes committed progenitors and plays a role in blast crisis transition in chronic myelogenous leukemia. Blood 115, 2872–2881 (2010)
Battelli, C., Nikopoulos, G. N., Mitchell, J. G. & Verdi, J. M. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1 . Mol. Cell. Neurosci. 31, 85–96 (2006)
Liu, G. et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer 5, 67 (2006)
Pece, S. et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167, 215–221 (2004)
Zhao, C. et al. Loss of β-catenin impairs the renewal of normal and CML stem cells in vivo . Cancer Cell 12, 528–541 (2007)
Han, H. et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14, 637–645 (2002)
Qin, X. F., An, D. S., Chen, I. S. & Baltimore, D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl Acad. Sci. USA 100, 183–188 (2003)
Jegga, A. G. et al. Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res. 12, 1408–1417 (2002)
Radich, J. P. et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc. Natl Acad. Sci. USA 103, 2794–2799 (2006)
Zhao, L. P., Prentice, R. & Breeden, L. Statistical modeling of large microarray data sets to identify stimulus-response profiles. Proc. Natl Acad. Sci. USA 98, 5631–5636 (2001)
Acknowledgements
We thank A. M. Pendergast, J. Chute, K. Itahana, L. Penalva and L. Grimes for advice and reagents; K.-i. Yamamura for the Msi2 gene-trap mice; T. Honjo for the Rbpj conditional mice; N. Gaiano for the TNR mice; D. Baltimore for the lentiviral shRNA constructs; and A. Means and B. Hogan for comments on the manuscript. We also thank M. Cook, B. Harvat and L. Martinek for cell sorting; M. Fereshteh for advice on analysis of patient samples; D. McDonnell and H. Wade for advice on ChIP experiments; S. W. Tian for help in collecting patient samples and A. Chen and S. Honeycutt for technical help. The BCR–ABL construct was a gift from W. Pear and the NUP98–HOXA9 construct a gift from G. Gilliland. T.I. is the recipient of a postdoctoral fellowship from the Astellas Foundation for Research on Metabolic Disorders, K.L.C. is the recipient of an American Heart Association predoctoral award, B.Z. received support from T32 GM007184-33 and T.R. is the recipient of a Leukemia and Lymphoma Society Scholar Award. This work was also supported by an LLS Translational Research grant and an ASH Junior Faculty Award to V.G.O., as well as NIH grants CA18029 to J.P.R., CA140371 to V.G.O., CA122206 to C.T.J. and DK63031, DK072234, AI067798, HL097767, DP1OD006430 and an Alexander and Margaret Stewart Fund grant to T.R. We are grateful for the support received from the Lisa Stafford Research Prize.
Author information
Authors and Affiliations
Contributions
T.I. and H.Y.K. designed the research, performed the majority of the experiments and helped write the paper. B.Z., K.L.C., J.B., W.E.L. and C.Z. provided experimental data and help; A.L. provided histopathological analysis; C.T.J., G.G., L.F., J.G., H.G., S.-H.K., D.-W.K. and C.C. provided human patient samples and experimental advice; T.I., H.Y.K., G.G. and B.Z. defined gene expression in patient samples by PCR; and V.G.O. and J.P.R. carried out all microarray and patient outcome analyses. T.R. conceived of the project, planned and guided the research, and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
T.I., H.Y.K., D.W.K., V.O., J.P.R. and T.R. are named inventors on a provisional patent application no. 61/178,370 titled ‘Diagnostic and treatment for chronic and acute myeloid leukemia’.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-8 with legends. (PDF 3249 kb)
Rights and permissions
About this article
Cite this article
Ito, T., Kwon, H., Zimdahl, B. et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature 466, 765–768 (2010). https://doi.org/10.1038/nature09171
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09171
This article is cited by
-
Asymmetric division of stem cells and its cancer relevance
Cell Regeneration (2024)
-
Down-regulation of Musashi-2 exerts antileukemic effects on acute lymphoblastic leukemia cells and increases sensitivity to dexamethasone
Annals of Hematology (2024)
-
MicroRNA-143 acts as a tumor suppressor through Musashi-2/DLL1/Notch1 and Musashi-2/Snail1/MMPs axes in acute myeloid leukemia
Journal of Translational Medicine (2023)
-
Prevalence, causes and impact of TP53-loss phenocopying events in human tumors
BMC Biology (2023)
-
Genetic deletion and pharmacologic inhibition of E3 ubiquitin ligase HOIP impairs the propagation of myeloid leukemia
Leukemia (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.