Abstract
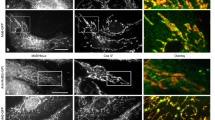
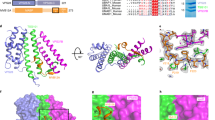
The interferon-inducible dynamin-like myxovirus resistance protein 1 (MxA; also called MX1) GTPase is a key mediator of cell-autonomous innate immunity against pathogens such as influenza viruses1. MxA partially localizes to COPI-positive membranes of the smooth endoplasmic reticulum–Golgi intermediate compartment2. At the point of infection, it redistributes to sites of viral replication and promotes missorting of essential viral constituents3,4. It has been proposed that the middle domain and the GTPase effector domain of dynamin-like GTPases constitute a stalk that mediates oligomerization and transmits conformational changes from the G domain to the target structure5,6,7; however, the molecular architecture of this stalk has remained elusive. Here we report the crystal structure of the stalk of human MxA, which folds into a four-helical bundle. This structure tightly oligomerizes in the crystal in a criss-cross pattern involving three distinct interfaces and one loop. Mutations in each of these interaction sites interfere with native assembly, oligomerization, membrane binding and antiviral activity of MxA. On the basis of these results, we propose a structural model for dynamin oligomerization and stimulated GTP hydrolysis that is consistent with previous structural predictions and has functional implications for all members of the dynamin family.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Haller, O., Stertz, S. & Kochs, G. The Mx GTPase family of interferon-induced antiviral proteins. Microbes Infect. 9, 1636–1643 (2007)
Stertz, S. et al. Interferon-induced, antiviral human MxA protein localizes to a distinct subcompartment of the smooth endoplasmic reticulum. J. Interferon Cytokine Res. 26, 650–660 (2006)
Kochs, G. & Haller, O. Interferon-induced human MxA GTPase blocks nuclear import of Thogoto virus nucleocapsids. Proc. Natl Acad. Sci. USA 96, 2082–2086 (1999)
Reichelt, M. et al. Missorting of LaCrosse virus nucleocapsid protein by the interferon-induced MxA GTPase involves smooth ER membranes. Traffic 5, 772–784 (2004)
Mears, J. A., Ray, P. & Hinshaw, J. E. A corkscrew model for dynamin constriction. Structure 15, 1190–1202 (2007)
Klockow, B. et al. The dynamin A ring complex: molecular organization and nucleotide-dependent conformational changes. EMBO J. 21, 240–250 (2002)
Chen, Y. J. et al. The stalk region of dynamin drives the constriction of dynamin tubes. Nature Struct. Mol. Biol. 11, 574–575 (2004)
Reubold, T. F. et al. Crystal structure of the GTPase domain of rat dynamin 1. Proc. Natl Acad. Sci. USA 102, 13093–13098 (2005)
Flohr, F. et al. The central interactive region of human MxA GTPase is involved in GTPase activation and interaction with viral target structures. FEBS Lett. 463, 24–28 (1999)
Arnheiter, H. & Haller, O. Antiviral state against influenza virus neutralized by microinjection of antibodies to interferon-induced Mx proteins. EMBO J. 7, 1315–1320 (1988)
Schwemmle, M. et al. Unexpected structural requirements for GTPase activity of the interferon-induced MxA protein. J. Biol. Chem. 270, 13518–13523 (1995)
Zurcher, T., Pavlovic, J. & Staeheli, P. Mechanism of human MxA protein action: variants with changed antiviral properties. EMBO J. 11, 1657–1661 (1992)
Chappie, J. S. et al. An intramolecular signaling element that modulates dynamin function in vitro and in vivo . Mol. Biol. Cell 20, 3561–3571 (2009)
Prakash, B. et al. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403, 567–571 (2000)
Daumke, O. et al. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature 449, 923–927 (2007)
Low, H. H. & Lowe, J. A bacterial dynamin-like protein. Nature 444, 766–769 (2006)
Hinshaw, J. E. & Schmid, S. L. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374, 190–192 (1995)
Ramachandran, R. et al. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 26, 559–566 (2007)
Ingerman, E. et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027 (2005)
Kochs, G. et al. Self-assembly of human MxA GTPase into highly ordered dynamin-like oligomers. J. Biol. Chem. 277, 14172–14176 (2002)
Ponten, A. et al. Dominant-negative mutants of human MxA protein: domains in the carboxy-terminal moiety are important for oligomerization and antiviral activity. J. Virol. 71, 2591–2599 (1997)
Accola, M. A. et al. The antiviral dynamin family member, MxA, tubulates lipids and localizes to the smooth endoplasmic reticulum. J. Biol. Chem. 277, 21829–21835 (2002)
Kochs, G. et al. Assay and functional analysis of dynamin-like Mx proteins. Methods Enzymol. 404, 632–643 (2005)
Richter, M. F. et al. Interferon-induced MxA protein. GTP binding and GTP hydrolysis properties. J. Biol. Chem. 270, 13512–13517 (1995)
Melen, K. et al. Enzymatic characterization of interferon-induced antiviral GTPases murine Mx1 and human MxA proteins. J. Biol. Chem. 269, 2009–2015 (1994)
Hefti, H. P. et al. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La crosse virus and other lethal viral infections. J. Virol. 73, 6984–6991 (1999)
Maines, T. R. et al. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 79, 11788–11800 (2005)
Dittmann, J. et al. Influenza A virus strains differ in sensitivity to the antiviral action of Mx-GTPase. J. Virol. 82, 3624–3631 (2008)
Bashkirov, P. V. et al. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell 135, 1276–1286 (2008)
Ghosh, A. et al. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature 440, 101–104 (2006)
Van Duyne, G. D. et al. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 (1993)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Pape, T. & Schneider, T. R. HKL2MAP: a graphical user interface for phasing with SHELX programs. J. Appl. Cryst. 37, 843–844 (2004)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 (1997)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Laskowski, R. A. et al. Procheck - a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26, 283–291 (1993)
Brünger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Kraulis, P. J. Molscript - a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24, 946–950 (1991)
Merritt, E. A. & Murphy, M. E. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr. D 50, 869–873 (1994)
Landau, M. et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 (2005)
Potterton, L. et al. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D 60, 2288–2294 (2004)
Guex, N. & Peitsch, M. C. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997)
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
DeLano, W. L. The PyMol Molecular Graphics System 〈http://www.pymol.org〉 (DeLano Scientific, 2002)
Steinkellner, G. et al. VASCo: computation and visualization of annotated protein surface contacts. BMC Bioinform. 10, 32 (2009)
Behlke, J., Ristau, O. & Schonfeld, H. J. Nucleotide-dependent complex formation between the Escherichia coli chaperonins GroEL and GroES studied under equilibrium conditions. Biochemistry 36, 5149–5156 (1997)
Praefcke, G. J. et al. Identification of residues in the human guanylate-binding protein 1 critical for nucleotide binding and cooperative GTP hydrolysis. J. Mol. Biol. 344, 257–269 (2004)
Acknowledgements
This project was supported by a grant of the Deutsche Forschungsgemeinschaft (SFB 740-From Molecules to Modules) and by a Career Development Fellowship of ‘The International Human Frontier Science Program Organization’ to O.D., and by the German-Israeli-Research foundation (GIF-841/04) to G.K. and O.H. We are grateful to J. Hinshaw and J. Mears for providing the EM maps and model fittings of oligomerized dynamin. We would also like to acknowledge help and support of O. Ristau and K. Schilling/Nanolytics (analytical ultracentrifugation), M. Dahte and H. Nikolenko (fluorescence measurements), A. Herrmann, T. Korte and P. Müller (stopped-flow analysis), G. Dittmar (mass spectrometry analysis), S. Werner and S. Gruber (technical assistance) and the BESSY staff at BL14.1 (data collection). This work was conducted by A.v.M. in partial fulfilment for a PhD degree from the Faculty of Biology at the University of Freiburg, Germany.
Author information
Authors and Affiliations
Contributions
S.G. solved the structure and carried out the biochemical characterization of MxA mutants. A.v.M. carried out all antiviral and cellular assays. S.P. assisted S.G. in cloning and purification. J.B. performed the analytical ultracentrifugation analysis. S.G., A.v.M., O.H., G.K. and O.D. planned the experimental design and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1-12 with legends, Supplementary Tables 1-2 and References. (PDF 3284 kb)
Supplementary Data
This file contains the Pdb coordinates (dyn-oligomer.pdb) of four dynamin molecules in our proposed oligomer. The stalks of MxA were aligned as described in Fig. 4 and Supp. Fig. 12. The G-domains are from rat Dynamin1 (pdb 2AKA, ref. 8) with the position of the GDP molecule derived from Dictyostelium Dynamin (pdb 1JWY, ref. 61). The coordinates of the PH domain are from pdb 2DYN (ref. 62). (TXT 1400 kb)
Rights and permissions
About this article
Cite this article
Gao, S., von der Malsburg, A., Paeschke, S. et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465, 502–506 (2010). https://doi.org/10.1038/nature08972
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08972
This article is cited by
-
Structural mechanism of mitochondrial membrane remodelling by human OPA1
Nature (2023)
-
Porcine interferon lambda 3 (IFN-λ3) shows potent anti-PRRSV activity in primary porcine alveolar macrophages (PAMs)
BMC Veterinary Research (2020)
-
PRV UL13 inhibits cGAS–STING-mediated IFN-β production by phosphorylating IRF3
Veterinary Research (2020)
-
Mx genes: host determinants controlling influenza virus infection and trans-species transmission
Human Genetics (2020)
-
Severe DNM1 encephalopathy with dysmyelination due to recurrent splice site pathogenic variant
Human Genetics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.