Abstract
Constitutive expression of telomerase in human cells prevents the onset of senescence and crisis by maintaining telomere homeostasis. However, accumulating evidence suggests that the human telomerase reverse transcriptase catalytic subunit (TERT) contributes to cell physiology independently of its ability to elongate telomeres. Here we show that TERT interacts with the RNA component of mitochondrial RNA processing endoribonuclease (RMRP), a gene that is mutated in the inherited pleiotropic syndrome cartilage–hair hypoplasia. Human TERT and RMRP form a distinct ribonucleoprotein complex that has RNA-dependent RNA polymerase (RdRP) activity and produces double-stranded RNAs that can be processed into small interfering RNA in a Dicer (also known as DICER1)-dependent manner. These observations identify a mammalian RdRP composed of TERT in complex with RMRP.
Similar content being viewed by others
Main
Telomerase is a ribonucleoprotein complex that elongates telomeres. Although several proteins interact with telomerase1,2,3,4, the minimal components of active telomerase include the catalytic telomerase reverse transcriptase (TERT) and a noncoding RNA (TERC) that encodes the template to synthesize telomeric DNA5. Telomere homeostasis mediated by telomerase maintains genomic stability and regulates cell lifespan6. Mutations in TERT, TERC or dyskerin, a telomerase-associated nucleolar protein involved in ribosomal RNA maturation7, are found in dyskeratosis congenita, a syndrome characterized by ectodermal dysplasia and bone marrow failure, and TERT mutations have been reported in aplastic anaemia and idiopathic pulmonary fibrosis8. Moreover, alterations in the regulation of telomeres and telomerase contribute to malignant transformation by affecting genomic integrity and cell immortalization6.
However, accumulating evidence suggests that TERT has activities beyond telomere maintenance9,10,11,12,13 and forms several intracellular complexes2,3,4. In particular, the overexpression of TERT induces increased tumour susceptibility9,10 and disrupts stem-cell function independently of telomere maintenance12, whereas the suppression of TERT expression alters global chromatin structure11. Indeed, some of these telomere-independent functions of TERT do not require the expression of TERC12.
Identification of a second RNA that interacts with TERT
To identify human TERT partners, we stably overexpressed a tandem affinity peptide (TAP)-tagged TERT protein in HeLa S3 cells, isolated TERT immune complexes, and identified a heterogeneous mixture of 38 RNA sequences associated with TERT (Supplementary Fig. 2 and Supplementary Table 1). We found that 5% of the sequences corresponded to TERC and the RNA component of mitochondrial RNA processing endoribonuclease (RMRP). RMRP is a 267-nucleotide noncoding RNA that is a small nucleolar RNA, like TERC, and is also found in mitochondria8,14. RMRP mutations are found in the pleiotropic inherited syndrome, cartilage–hair hypoplasia15.
From a single immune complex, we confirmed that either overexpressed or endogenous TERT interacts with RMRP and TERC, by isolating TAP–TERT (Fig. 1a) or endogenous TERT (Fig. 1b) complexes in both HeLa and 293T cells under conditions in which we failed to recover the ribozyme RNase P. We also found that the abundance of TERT–RMRP and TERT–TERC complexes was similar, even though TERC was expressed at five-fold higher levels than RMRP in these cells (Fig. 1c and Supplementary Fig. 3).
a, Detection of RMRP and TERC. RNA species associated with TAP–TERT complexes from a single immunoprecipitation (IP) were isolated and subjected to PCR with reverse transcription (RT–PCR). RT (-) indicates the absence of reverse transcriptase. Right panel shows the levels of TAP–TERT. HA, haemagglutinin; IB, immunoblot. b, TERT interacts with endogenous RMRP. TERT complexes from 293T and HeLa cells were isolated with an anti-TERT antibody and associated RNAs were subjected to RT–PCR. c, RNAs purified from TERT complexes isolated from HeLa S3 cells expressing TAP–TERT or a control vector or 293T cells were subjected to northern blotting. Ab, antibody.
To characterize the interaction between TERT and RMRP, we used TERT truncation mutants and found that the amino terminal end of TERT (1–531) was necessary for interactions with RMRP (Supplementary Fig. 4). This region overlaps with two regions required for the binding of TERC8,16. These observations demonstrate that TERT and RMRP form a new ribonucleoprotein complex distinct from the TERT–TERC enzyme.
The TERT–RMRP complex has RdRP activity
To test whether RMRP substitutes for TERC to reconstitute telomerase activity, we combined recombinant TERT with TERC or RMRP RNAs transcribed in vitro. Although we detected telomerase activity with TERT and TERC (Supplementary Fig. 5), we failed to detect telomerase activity when TERT and RMRP were co-incubated.
TERT has also been shown to act as a terminal transferase17, and human TERT shares sequence similarity to both viral reverse transcriptases and RdRPs18. RdRPs participate in the endogenous RNA interference (RNAi) pathway and in the regulation of post-transcriptional gene silencing19,20,21,22,23. To examine whether the TERT–RMRP complex has RdRP and/or terminal transferase activity, we established an RNA synthesis activity assay with recombinant TERT protein (Supplementary Fig. 6) and RNA molecules transcribed in vitro. We predicted three modes that the TERT–RMRP complex might use to elongate RNA: (1) as an RdRP that uses a de-novo-synthesized RNA primer to elongate a complementary strand (Fig. 2a, left panel); (2) as an RdRP that uses a 3′ fold-back (back-priming) configuration of template RNA as a primer (Fig. 2a, middle panel); or (3) as a terminal transferase (Fig. 2a, right panel). Viral RdRPs24,25 have been shown to use the first two modes to prime RdRP activity, and cellular RdRPs in fission yeast26 and fungi23 use similar priming mechanisms to produce double-stranded (ds) RNAs that act as precursors for RNAi.
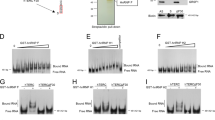
a, Predicted RNA products produced by RdRP or terminal transferase (TT) activity. b, RNA products produced by the RdRP activity derived from recombinant TERT and RMRP. nt, nucleotides. c, d, Treatment of RNA products with RNase T1 (c) or bacterial RNase III (d). e, RdRP assay performed in the presence of ribonucleotides (middle) or in the absence of adenine (left lane) or guanine (right lane) ribonucleotides. A and G are present within the first 5 nucleotides of the predicted complementary strand of RMRP. f, TERT-DN binds RMRP but lacks RdRP activity. TERT immune complexes were isolated from 293T cells expressing Flag-tagged TERT or Flag-tagged TERT-DN. RdRP activity is shown in the bottom panel. WT, wild-type. g, Northern blotting to detect complementary sequence of RMRP. h, Time course of RdRP activity. i, RNA products produced by recombinant TERT and truncation mutants of RMRP transcribed in vitro. Faint signals at 200, 120 and 60 nucleotides are TERT terminal transferase products. j, RNA products produced by the RdRP activity derived from recombinant TERT or TERT-DN and total RNA. A limited pool of RNAs serves as template for RdRP activity.
We found that recombinant TERT and RMRP produced two different products depending on the salt concentration (Fig. 2b and Supplementary Fig. 7). Specifically, we found ∼267-nucleotide- (corresponding to sense RMRP) and ∼534-nucleotide-sized products (hereafter referred to as sense plus antisense RMRP products) under high salt conditions, and RMRP-sized products under low salt conditions. To discriminate between these modes, we treated the products of the RdRP assay with RNase T1 (Fig. 2c) using conditions that favour the digestion of single-stranded RNA. RNase T1 treatment eliminated the ∼267-nucleotide RMRP-sized RNA products produced under low salt concentrations (data not shown), indicating that [32P]UTP was incorporated by terminal transferase activity.
In contrast, under high salt conditions, we found two RNAs (∼267 and ∼534 nucleotides) that collapsed into a single ∼267-nucleotide band after treatment with RNase T1 (Fig. 2c). To eliminate the possibility that the sense plus antisense product represented partially denatured RNAs, we treated the products of the RdRP assay with bacterial RNase III to digest dsRNA, and found that only the input ∼267-nucleotide RNA remained (Fig. 2d). Furthermore, when we left out adenine or guanine ribonucleotides, we failed to detect the sense plus antisense product (Fig. 2e). These observations confirm that the ∼534-nucleotide sense plus antisense products are formed by RdRP activity and represent a double-stranded hairpin structure created by an RNA molecule composed of sense and antisense strands of RMRP.
To confirm that the interaction between TERT and RMRP was required for RdRP activity, we performed an RdRP activity assay using combinations of recombinant mutant TERT proteins and RMRP. We failed to detect RdRP reaction products when TERT and TERC were co-incubated (Supplementary Fig. 8). Moreover, when we used the TERT-HT1 mutant that does not bind RMRP (Supplementary Fig. 4), we failed to observe labelled RNA products (Supplementary Fig. 8) under conditions in which we detected two different RNA products in reactions containing wild-type TERT and RMRP. We previously described a catalytically inactive TERT mutant (TERT-DN) that fails to elongate telomeres11,27. We confirmed that the recombinant TERT-DN mutant retained the ability to bind RMRP (Fig. 2f), but that the TERT-DN–RMRP complex lacked detectable RdRP activity (Fig. 2f). Thus TERT acts as the catalytic subunit for both the telomerase reverse transcriptase and RdRP activities.
TERT–RMRP RdRP produces dsRNA
These observations suggest that the TERT–RMRP RdRP synthesizes dsRNA in a template-dependent manner. To confirm the synthesis of the RMRP complementary strand, we used the sense strand of RMRP as a probe in northern blotting. We detected the antisense strand of RMRP in reactions containing recombinant wild-type TERT protein and RMRP, but not in reactions containing TERT-DN and RMRP (Fig. 2g). Furthermore, we detected the sense plus antisense product in the RdRP assay using the antisense strand of RMRP as a probe (Supplementary Fig. 9). These observations indicate that the TERT–RMRP RdRP produces dsRNAs in a template-dependent manner in vitro.
To determine whether the TERT–RMRP RdRP uses a back-priming mechanism, we examined the priming process using TERT and RMRP as a model system and found that elongation products appeared in a time-dependent manner (Fig. 2h and Supplementary Fig. 10). To assess whether the RMRP RNA forms a 3′ fold-back configuration, we generated 3′ RMRP truncation mutants and failed to find any reaction products (Fig. 2i). Thus, unlike what has been described for other cellular RdRPs, the TERT–RMRP RdRP has a restricted preference for RNA molecules that can be used as a template. Indeed, when we incubated purified recombinant TERT together with total cellular RNA and [32P]UTP, we identified a limited number of labelled RNAs (Fig. 2j). Although the secondary structure adopted by RMRP to create the 3′ fold-back is not known, these observations suggest that RMRP can itself serve as a primer for the polymerization process using a 3′ fold-back structure.
To ascertain whether this RdRP activity also occurs in vivo, we used the sense strand of RMRP as a probe and found ∼534-nucleotide RNAs that contain antisense RMRP in RNA derived from 293T, HeLa and MCF7 cells (Fig. 3a and Supplementary Figs 11 and 12). Moreover, we detected sense products and sense plus antisense products using RMRP antisense-strand probe (Fig. 3b). These observations confirmed that the ∼534-nucleotide products contain both sense and antisense RMRP sequences. To determine whether TERT was necessary for the appearance of antisense RMRP in cells, we examined the levels of the complementary RMRP strand in cells that do not express TERT and TERC (VA-13 cells)28, in cells that transiently express low levels of TERT (BJ cells)27,29,30, and in cells that constitutively express TERT (293T and HeLa cells). We also introduced a control vector or a vector that encodes TERT in VA-13 and BJ cells. We detected the complementary RMRP strand using a quantitative RNase protection assay with a sense-strand probe that detects antisense RMRP (Fig. 3c and Supplementary Fig. 13), and using northern blotting with both sense and antisense strand-specific RMRP probes (Fig. 3d and Supplementary Fig. 11a). The levels of antisense RMRP correlated with the expression of TERT (Fig. 3c, d). These observations confirmed that the TERT–RMRP RdRP produces double-stranded RMRP in vivo.
a, Northern blotting to detect complementary sequence of RMRP in cell lines. ‘+’ indicates samples treated with RNase. b, Northern blotting to detect the RMRP sense strand. c, TERT expression correlates with the levels of antisense (AS) RMRP detected by RNase protection assay. Vector denotes cells infected with a control vector. d, TERT expression correlates with the levels of the sense (S) plus antisense RMRP products detected by northern blotting. The bottom panel shows levels of the small nuclear RNA U2.
Effects of the TERT–RMRP complex on RMRP expression
To assess the consequences of overexpressing the TERT–RMRP complex on RMRP levels, we introduced RMRP into cells that lack TERT expression (VA-13), that transiently express TERT in a cell-cycle-dependent manner (BJ fibroblasts), and that constitutively express TERT (VA-13 and BJ fibroblasts expressing ectopic TERT, and HeLa and MCF7 cells). After expressing RMRP in cells lacking TERT (VA-13), we found that RMRP levels were increased (Fig. 4a and Supplementary Fig. 14). In contrast, in cells that express TERT, we found that the steady-state levels of RMRP were decreased when RMRP was overexpressed, regardless of the promoter that was used to express RMRP (Fig. 4a and Supplementary Fig. 14). We also found that forced TERT expression in VA-13 or BJ cells suppressed RMRP expression (Fig. 4b and Supplementary Fig. 15). Consistent with these findings, suppression of TERT in HeLa cells led to increased RMRP expression (Fig. 4c).
a, Semi-quantitative RT–PCR for total RMRP and retrovirally delivered RMRP (ectopic) in cell lines expressing control or RMRP expression vectors. Promoters used to express RMRP are indicated. The relative intensity of RMRP is noted below each panel. CMV, cytomegalovirus. See Supplementary Fig. 14. b, RT–PCR for total RMRP. See Supplementary Fig. 15. c, Effects of suppressing TERT on RMRP levels. A control shRNA (green fluorescent protein (GFP) shRNA) or two different TERT-specific shRNAs were stably introduced into HeLa cells. d, Effects of RMRP mutants on RMRP levels. LTR, long terminal repeat. RT–PCR was used to detect RMRP levels in c and d. e, Detection of small RNA species in human cells. Northern blotting to detect small RNAs (22 nucleotides in length) using antisense (left panel) and sense (right panel) probes derived from nucleotides 21–40 of RMRP. f, g, Analysis of the termini of the small RNA species identified in e. Total RNA was incubated with the indicated enzyme (f), or oxidation-β-elimination reactions (g) were performed. Northern blotting was performed with antisense probe. CIP, calf intestinal phosphatase; PNK, polynucleotide kinase. ATP- indicates samples lacking ATP.
Because the 3′ end of RMRP was essential for TERT–RMRP activity (Fig. 2i), we examined the effects of expressing RMRP truncation mutants lacking 3′ ends and found that only truncation mutants lacking intact 3′ ends were readily overexpressed (Fig. 4d). These observations demonstrate that RMRP expression levels are dependent on the TERT–RMRP RdRP and suggest that RMRP levels are controlled by an RdRP-dependent, negative-feedback mechanism.
Identification of siRNAs derived from RMRP
In other organisms, RdRPs synthesize dsRNAs that are processed into active short interfering RNAs (siRNAs)31. Because manipulating TERT and RMRP levels affected RMRP expression, we proposed that the TERT–RMRP complex produces RMRP-specific siRNA to regulate RMRP levels. To test this possibility, we used sense and antisense probes corresponding to RMRP (nucleotides 21–40) in northern blotting and found double-stranded 22-nucleotide RNAs (Fig. 4e and Supplementary Fig. 11b). Because siRNAs contain 5′ monophosphate and 3′ hydroxyl groups32,33,34, we characterized the chemical nature of the small RNA ends. We found that calf intestinal phosphatase slowed the migration of these short RNAs, and subsequent incubation with polynucleotide kinase and ATP restored the mobility of the short RNAs, indicating that either the 5′ or the 3′ end of this small RNA is monophosphorylated (Fig. 4f and data not shown). Moreover, incubation with polynucleotide kinase in the absence of ATP did not alter the migration (Fig. 4f), and oxidation and β-elimination treatment increased the migration of these small RNAs (Fig. 4g), indicating that the 3′ ends bear vicinal 2′,3′ dihydroxyls. Together, these observations confirm that these small RNAs contain 5′ monophosphate and 3′ hydroxyl groups, and therefore share the size and chemical composition of known siRNAs.
To demonstrate that dsRNAs produced by the TERT–RMRP RdRP are processed into siRNA, we suppressed the expression of Dicer with two distinct Dicer-specific short hairpin RNAs (shRNAs). Suppression of Dicer to levels that partially inhibited the processing of the microRNA miR-16 (Fig. 5a and Supplementary Fig. 16) led to diminished levels of the siRNAs derived from RMRP (Fig. 5a). When we suppressed Dicer expression in HeLa, 293T or MCF7 cells, we found that endogenous RMRP levels increased up to 3.7-fold (Fig. 5b). Suppressing Dicer expression in VA-13 cells that lack TERT did not affect the levels of single-stranded RMRP (Fig. 5b), but did increase levels of the elongated sense plus antisense RMRP products in cells that constitutively express TERT (Supplementary Fig. 17). Moreover, we found that only the sense strands of these endogenous RMRP-specific siRNAs were associated with human AGO2 (also known as EIF2C2; Fig. 5c). These observations indicate that the endogenous RMRP-specific siRNAs are processed by the RNA-induced silencing complex, similar to other small RNAs that are processed into siRNA.
a, Effect of suppressing Dicer on RMRP-derived small RNAs. Northern blotting was performed to detect: (1) small RNAs using the antisense strand of RMRP as a probe in the indicated cells expressing control shRNA (GFP shRNA) or Dicer-specific shRNAs (Dicer shRNA1 and shRNA2); (2) precursor microRNA pre-miR-16 and mature miR-16 using a miR-16-specific probe; and (3) U6 RNA. See Supplementary Fig. 16. b, RT–PCR for total RMRP from cell lines expressing control shRNA or Dicer-specific shRNAs. IB, immunoblot. The relative intensity of RMRP is noted at the bottom of the panel. c, RMRP-derived small RNAs are associated with AGO2. Human AGO2 immune complexes were isolated using anti-AGO2-specific antisera or pre-immune sera, and small RNAs were detected by northern blotting. Blotting of oligonucleotides (RMRP 20–41 and RMRP AS 41–20) is also shown.
To confirm that these small RNAs act as siRNAs, we identified small RNAs from total RNA that hybridized to probes spanning RMRP, synthesized siRNA corresponding to the identified sequences, and tested the consequences of introducing this siRNA in HeLa, 293T and MCF7 cells. We found that the synthesized siRNA suppressed endogenous RMRP levels (Supplementary Fig. 18). These observations provide evidence that similar to other cellular RdRPs, the TERT–RMRP RdRP synthesizes dsRNAs that act as a precursor for siRNAs.
Discussion
Here we demonstrate that human TERT and RMRP form a distinct ribonucleoprotein complex that has the ability to produce dsRNAs (Supplementary Fig. 1). Like RdRPs found in other organisms, the human TERT–RMRP complex produces dsRNAs that act as substrates for the generation of siRNA. However, unlike other cellular RdRPs23,26,31,35,36, the human TERT–RMRP RdRP shows a strong preference for RNA templates that can form 3′ fold-back structures. Because other cellular RdRPs have been identified using assays that require primer-independent RdRP activity23,26,36, the substrate specificity of the human TERT–RMRP RdRP may, in part, account for the difficulty in identifying mammalian enzymes that have RdRP activity.
Although the cellular RdRPs described until now do not show a primer requirement, several viral RdRPs use both primer-dependent and primer-independent mechanisms, and fungal and yeast RdRPs are also able to use a back-priming mechanism23,26. Because TERT is a closed right-handed polymerase37 evolutionarily related to both reverse transcriptases and viral RdRPs18, these observations are consistent with previous observations that indicate that right-handed RdRPs exhibit primer-dependent RdRP polymerase activity38.
Using RMRP as a template, the TERT–RMRP RdRP produces dsRNAs that are processed by Dicer into 22-nucleotide dsRNAs that contain 5′ monophosphate and 3′ hydroxyl groups and are loaded into AGO2, confirming that these short RNAs represent endogenous siRNAs. Recent work has shown that in oocytes and embryonic stem cells, endogenous siRNA can also be formed by the transcription of complementary sense and antisense strands39,40,41. Thus, in mammals at least two mechanisms lead to the production of dsRNAs that are processed into siRNA. Further work will be necessary to determine whether there are tissue-dependent differences in the use of these two mechanisms and whether other mammalian RdRPs exist.
We found that the TERT–RMRP RdRP regulates RMRP levels by a negative-feedback control mechanism. The identities and functions of the RNAs other than RMRP that act as templates for the TERT–RMRP RdRP remain to be identified (Fig. 2j). However, because endogenously encoded siRNAs suppress L1 retrotransposition in human cells42, these observations suggest that the TERT–RMRP complex may regulate the expression of other genes by generating siRNAs.
Because mutations in RMRP are found in cartilage–hair hypoplasia15, these findings suggest that perturbation of the TERT–RMRP complex is involved in the pathogenesis of this disorder. The involvement of human TERT in two syndromes characterized by stem-cell failure (cartilage–hair hypoplasia and dyskeratosis congenita)7,8,43 suggests that ribonucleoprotein complexes containing TERT has a critical role in stem cell biology. Indeed, overexpression of mouse TERT in mice lacking Terc leads to defects in normal hair follicle stem-cell function12 at least in part by altering gene expression programs related to stem cell function44. In mammals, TERT may regulate both telomere biology and gene expression through these two ribonucleoprotein complexes.
Methods Summary
RNAs that bind TERT were identified from HeLa S3 cells expressing a TAP epitope-tagged TERT. RNAs that bound to TERT after two rounds of purification were analysed using an Experion capillary electrophoresis device (Bio-Rad) to visualize RNA species. For RNA cloning and sequencing, the same samples were separated using a 7 M urea/15% polyacrylamide gel, and RNAs recovered from the gel were cloned using a small RNA cloning Kit (TaKaRa). Purified glutathione S-transferase (GST)–TERT was isolated from Escherichia coli and incubated with either TERC or RMRP transcribed in vitro, to assess the ability of such complexes to exhibit telomerase or RdRP activity. RNAi was used to suppress TERT and to show that the TERT–RMRP complex also produces dsRNA in cells. Northern blotting with sense and antisense probes specific for RMRP (nucleotides 21–40) identified 22-nucleotide, double-stranded RNAs that contained a 5′ monophosphate and a 3′ hydroxyl group, which were loaded into human AGO2. To determine the function of these RMRP-derived small RNAs, a chemically synthesized siRNA corresponding to these small RNAs (siRNA: 5′-GGCTACACACTGAGGACTC-3′; Dharmacon) was transfected into HeLa, 293T and MCF7 cells.
Online Methods
Cell culture and stable expression of TAP–TERT
The human cell lines 293T, MCF7, HeLa, HeLa S3 and VA-13 were maintained in DMEM supplemented with 10% heat-inactivated FBS. BJ fibroblasts were cultured as described45. Amphotropic retroviruses were created as described45,46 using the vectors pWZL-Blast-N-Flag/HA-TERT (for HeLa-S3-TAP–TERT), pBABE-puro or pBABE-puro-TERT. After infection, cells were selected with blasticidin S (10 μg ml-1) for 5 days or with puromycin (2 μg ml-1) for 3 days.
Purification of TERT complexes and cloning of RNAs
HeLa S3 cells (2 × 108) expressing or lacking (control) TAP–TERT were lysed in 5 ml of lysis buffer A (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% NP-40, 0.1 mM dithiothreitol (DTT)) and incubated for 30 min on ice. The lysate was then pelleted by centrifugation (16,000g) for 20 min at 4 °C. The supernatant was incubated with anti-Flag (M2) antibody-conjugated agarose overnight at 4 °C. The beads were washed three times with lysis buffer A and eluted with 3× Flag peptide (150 ng μl-1). The resulting elution was incubated with Protein A Sepharose beads and an anti-HA antibody (F7; Santa Cruz) for 4 h at 4 °C. The beads were washed three times with lysis buffer A, and RNA was isolated using TRIzol (Invitrogen). RNA samples prepared in this manner were analysed using an Experion capillary electrophoresis device (Bio-Rad) to visualize RNA species. For RNA cloning and the sequencing, the same samples were separated using a 7 M urea/15% polyacrylamide gel, and RNAs recovered from gel were cloned using a small RNA cloning Kit (TaKaRa).
RNA preparation for immunoprecipitation RT–PCR
RNA samples that were prepared from the HeLa S3 cells expressing TAP–TERT as described earlier were also subjected to RT–PCR. For immunoprecipitation of endogenous TERT complexes, cells (1 × 108) were lysed in 600 μl of lysis buffer A, sonicated and pre-cleared with 15 μl of 50% slurry of Protein A Sepharose (Pierce) for 2 h at 4 °C. The pre-cleared total cell lysate was incubated with a rabbit polyclonal anti-TERT antibody (Rockland, 2 μl) for 3 h at 4 °C, followed by incubation with 30 μl of 50% slurry of Protein A Sepharose overnight at 4 °C. After binding, the beads were washed three times for 30 min with lysis buffer A. RNA derived from a single immunoprecipitation was isolated from the Protein A Sepharose using TRIzol (Invitrogen) followed by RT–PCR with primers specific for TERC, RMRP or RNase P. Although other RNAs also co-purified with human TERT (Supplementary Table 1), we failed to confirm the interaction of Alu sequences or the 5.8S ribosomal RNA on the Y chromosome with TERT (data not shown).
RT–PCR and quantitative RT–PCR
Either total cellular RNA or RNA from immunoprecipitation was isolated using TRIzol (Invitrogen) and subjected to RT–PCR. The following primers were used: TERC (43F, 5′-TCTAACCCTAACTGAGAAGGGCGT-3′ and 163R, 5′-TGCTCTAGAATGAACGGTGGAAGG-3′), RMRP (F5, 5′-TGCTGAAGGCCTGTATCCT-3′ and R257, 5′-TGAGAATGAGCCCCGTGT-3′), RNase P (F50, 5′-GTCACTCCACTCCCATGTCC-3′ and R318, 5′-AATTGGGTTATGAGGTCCC-3′), and the human β-actin gene (also known as ACTB) (5′-CAAGAGATGGCCACGGCTGCT-3′ and 5′-TCCTTCTGCATCCTGTCGGCA-3′). The reverse transcription reaction was performed for 60 min at 42 °C using the recovered RNA, and PCR was immediately performed (22 cycles for 293T cells, and 26 cycles for HeLa cells: 94 °C, 30 s; 60 °C, 30 s; 72 °C, 30 s).
Quantitative RT–PCR (qRT–PCR) was performed with a LightCycler 480 II (Roche) according to the manufacturer’s protocols. The expression levels of RMRP were detected using the following primers and probe; forward primer, 5′-GAGAGTGCCACGTGCATACG-3′, reverse primer, 5′-CTCAGCGGGATACGCTTCTT-3′, VIC-labelled TaqMan MGB probe, 5′-ACGTAGACATTCCCC-3′. β-actin was used as a reference.
Total RMRP was detected using primers (F5, 5′-TGCTGAAGGCCTGTATCCT-3′ and R257, 5′-TGAGAATGAGCCCCGTGT-3′) that amplify both endogenous and ectopically introduced RMRP. In Fig. 4a, for VA-13, BJ and MCF7 cells, reverse transcription was performed using random hexamers (GE Healthcare) and ectopically expressed RMRP was detected with vector-specific primers (F5, 5′-TGCTGAAGGCCTGTATCCT-3′ and LKO.1-RT, 5′-ACTGCCATTTGTCTCGAGGT-3′). For HeLa cells, reverse transcription was performed with pQC3′ (5′-AAGCGGCTTCGGCCAGTAACGTTA-3′) and PCR was performed with the primers F5 (5′-TGCTGAAGGCCTGTATCCT-3′) and R257 (5′-TGAGAATGAGCCCCGTGT-3′). Northern blotting and qRT–PCR experiments (Supplementary Fig. 14) confirmed that the differences in RMRP levels that were observed using the RT–PCR conditions used in Fig. 4a accurately reflect RMRP levels. Signal intensity was measured with ImageJ software.
Telomerase activity reconstituted in vitro and TRAP assay
In vitro reconstitution of telomerase activity (telomere-specific reverse transcriptase activity) was performed as described previously5. In brief, recombinant TERT was expressed in the TnT T7-Coupled Reticulocyte Lysate System (Promega) following the manufacturer’s instructions. Purified TERC or RMRP was included in the in vitro transcription/translation reactions. The telomeric repeat amplification protocol (TRAP)45,46,47 was used to detect telomere-specific reverse transcriptase activity.
Affinity purification of recombinant GST–TERT fusion proteins
GST–TERT–HA, GST–TERT-HT1 and GST–TERT-DN in the pGENKZ expression vector48 were provided by S. Murakami. Bacteria (BL21-Gold) containing these vectors were plated at 30 °C overnight and then a single colony was picked to inoculate liquid cultures, which were incubated at 37 °C overnight. Thereafter 1 ml of this culture was re-inoculated into 100 ml of Luria-Bertani medium, incubated at 37 °C for 4 h without isopropyl-β-d-thiogalactoside (IPTG) induction, collected by centrifugation, suspended in a lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% NP-40, 0.1 mM DTT, 10 mM phenylmethyl sulphonyl fluoride (PMSF), proteinase inhibitor (Nacalai Tesque)) and sonicated twice for 10 s at 4 °C. After centrifugation of the sonicated lysates, the supernatants were passed through DEAE-Sepharose, and the GST-fusion proteins were recovered using glutathione-Sepharose 4B beads. The resin was washed with lysis buffer A at least three times, and the GST-fusion proteins were then eluted with glutathione at 4 °C for 1 h (20 mM glutathione (reduced form)) in elution buffer (50 mM Tris-HCl, pH 8.8, 150 mM NaCl, 0.5% NP-40, 0.1 mM DTT, 10 mM PMSF, proteinase inhibitor (Nacalai Tesque)). Supplementary Fig. 6 shows that wild type and TERT-DN were produced at similar levels using this method and the effects of incubation time and IPTG on yield. The average yield for this method is 500 ng (5 ng μl-1) of active form of TERT from 100 ml culture.
RdRP assay
The affinity purified recombinant GST–TERT fusion protein (10 ng) was incubated with 1 μg of full length RMRP RNA or truncated RMRP products (RMRP 1–200, RMRP 1–120 and RMRP 1–60 for Fig. 2i) transcribed in vitro (SP6) in 200 mM KCl, 50 mM Tris-HCl, pH 8.3, 10 mM DTT, 30 mM MgCl2, 50 μM rATP, 50 μM rGTP, 50 μM rCTP and 2 μCi of [α-32P]UTP at 32 °C for 2 h. To perform the experiments under low salt conditions, 20 μl of 0.2× SSC was then added to adjust final salt concentration to 15 mM NaCl and 1.5 mM sodium citrate, whereas 20 μl of 4× SSC was added to adjust final salt concentration to 300 mM NaCl and 30 mM sodium citrate to achieve high salt conditions. These mixtures were incubated at 37 °C for a further 1 h. Resulting products were treated with proteinase K to stop the reaction and purified with phenol–chloroform. To ensure that RNA products were completely denatured, we performed both conventional formamide treatment (with 95% formamide/20 mM EDTA gel-loading buffer at 95 °C for 5 min) and a further treatment with 1 M de-ionized glyoxal at 65 °C for 15 min.
To analyse double-stranded RNA produced by the TERT–RMRP complex, we performed this RdRP assay and treated the products with bacterial RNase III (E. coli, Ambion; 50 mM NaCl, 10 mM Tris-HCl, pH 7.9, 1 mM DTT, 10 mM MgCl2) or RNase T1 (Roche; 50 mM Tris-HCl, pH 8.3, 300 mM NaCl and 30 mM sodium citrate).
Northern blotting
Total RNA and small RNAs (<200 nucleotides in length) were isolated using a mirVana miRNA Isolation Kit (Ambion) according to the manufacturer’s protocol. Total RNA or small RNA (10 μg) was separated on denaturing polyacrylamide gels, then blotted onto Hybond-N+ membranes (GE Healthcare) using a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). Hybridization was performed in Church buffer (0.5 M NaHPO4, pH 7.2, 1 mM EDTA and 7% SDS) containing 106 c.p.m. ml-1 of each 32P-labelled probe for 14 h. The membranes were washed in 2× SSC, and the signals were detected by autoradiography.
Identification of short RNA species derived from RMRP
Using ten consecutive probes corresponding to the RMRP sequence, we found that the small RNAs derived from RMRP shown in Figs 4e–g and 5a were detected by probes containing the complementary sequences to nucleotides 21–40 of RMRP. To determine the function of these RMRP-derived small RNAs, we purchased a chemically synthesized siRNA targeting this 20-nucleotide portion of the RMRP sequence (siRNA: 5′-GGCTACACACTGAGGACTC-3′; Dharmacon) and transfected this siRNA into HeLa, 293T and MCF7 cells plated on six-well dishes using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
RNase protection assay
RMRP RNA was transcribed with SP6 RNA polymerase in the presence of [α-32P]UTP using RiboMAX Large Scale RNA Production System (Promega). Total cellular RNA (30 μg) was hybridized overnight at 60 °C with equal amounts of 32P-labelled RMRP sense probe. Hybrids were digested with RNase A and RNase T1. The protected fragments were separated by PAGE under denaturing conditions and visualized by autoradiography.
Analysis of the chemical structure of the ends of small RNAs
To determine the phosphorylation status of the termini of small RNAs, 30 μg of small RNA (<200 nucleotides in length) was treated with calf intestinal alkaline phosphatase (CIP; TaKaRa) for 2 h at 37 °C. CIP was inactivated by phenol–chloroform extraction. Part of the CIP-treated RNA was then treated with T4 polynucleotide kinase (TaKaRa) supplemented with 1 mM ATP for 2 h at 37 °C, and phenol–chloroform extraction was performed. Small RNA (15 μg) was treated with T4 polynucleotide kinase without ATP for 2 h at 37 °C. The reaction was inactivated by phenol–chloroform extraction. After overnight sodium acetate–ethanol precipitation at -20 °C, the treated RNAs were resolved by 20% denaturing polyacrylamide/urea gel electrophoresis and then analysed by northern blotting42,43.
To further analyse the 3′ end of these small RNAs, we performed oxidation and β-elimination reactions. Specifically, the NaIO4 reaction was performed by adding 20 μg of small RNA in water to 5× borate buffer (148 mM borax and 148 mM boric acid, pH 8.6) and freshly dissolved 200 mM NaIO4 to create a final concentration of 1× borate buffer and 25 mM NaIO4. The mixtures were incubated for 10 min at 20 °C. Glycerol was added to quench remaining NaIO4, and the samples were incubated for a further 10 min at 20 °C. For β-elimination, small RNAs were dried by centrifugation and evaporation and dissolved in 50 μl of 1× borax buffer (30 mM borax, 30 mM boric acid and 50 mM NaOH, pH 9.5) and incubated at 45 °C for 90 min. Nucleic acids were recovered by sodium acetate–ethanol precipitation at -20 °C overnight, and the treated RNAs were resolved by 20% denaturing 7 M urea PAGE and analysed by northern blotting43.
Stable expression of shRNA
We used the pLKO.1-puro vector and the sequences described below to create shRNA vectors specific for TERT, Dicer and GFP. These vectors were used to make amphotropic retroviruses and polyclonal cell populations were purified with selection with puromycin (2 μg ml-1). The sequences used for the indicated short hairpin RNAs are shown below where the capitalized letters represent the targeting sequences: TERT shRNA1, 5′-GGAAGACAGTGGTGAACTTCCctcgagGGAAGTTCACCACTGTCTTCCttttt-3′ and 5′-aattcaaaaaGGAAGACAGTGGTGAACTTCCctcgagGGAAGTTCACCACTGTCTTCC-3′; TERT shRNA2, 5′-GGAACACCAAGAAGTTCATCTctcgagAGATGAACTTCTTGGTGTTCCttttt-3′ and 5′-aattcaaaaaGGAACACCAAGAAGTTCATCTctcgagAGATGAACTTCTTGGTGTTCC-3′. Dicer sequences: Dicer shRNA1, 5′-GCTCGAAATCTTACGCAAATActcgagTATTTGCGTAAGATTTCGAGCtttttg-3′ and 5′-aattcaaaaaGCTCGAAATCTTACGCAAATActcgagTATTTGCGTAAGATTTCGAGC-3′; Dicer shRNA2, 5′-CCACACATCTTCAAGACTTAActcgagTTAAGTCTTGAAGATGTGTGGtttttg-3′ and 5′-aattcaaaaaCCACACATCTTCAAGACTTAActcgagTTAAGTCTTGAAGATGTGTGG-3′.
Immunoprecipitation of human AGO2 complexes
HeLa or 293T cells were lysed in lysis buffer A and immunoprecipitation was performed using pre-immune sera or anti-AGO2 antibodies49 (provided by H. Siomi and M. C. Siomi). RNA was isolated using TRIzol from the protein A beads and resolved by electrophoresis on 7 M urea 20% PAGE. Small RNAs were detected by northern blotting with an antisense probe, a sense probe derived from nucleotides 21–40 of RMRP, or a miR-16-specific probe (5′-CGCCAATATTTACGTGCTGCTA-3′).
References
Cohen, S. B. et al. Protein composition of catalytically active human telomerase from immortal cells. Science 315, 1850–1853 (2007)
Fu, D. & Collins, K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol. Cell 28, 773–785 (2007)
Venteicher, A. S., Meng, Z., Mason, P. J., Veenstra, T. D. & Artandi, S. E. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132, 945–971 (2008)
Venteicher, A. S. et al. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 323, 644–648 (2009)
Weinrich, S. L. et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet. 17, 498–502 (1997)
Chan, S. W. & Blackburn, E. H. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene 21, 553–563 (2002)
Liu, J. M. & Ellis, S. R. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood 107, 4583–4588 (2006)
Calado, R. T. & Young, N. S. Telomere maintenance and human bone marrow failure. Blood 111, 4446–4455 (2008)
Gonzalez-Suarez, E. et al. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. EMBO J. 20, 2619–2630 (2001)
Artandi, S. E. et al. Constitutive telomerase expression promotes mammary carcinomas in aging mice. Proc. Natl Acad. Sci. USA 99, 8191–8196 (2002)
Masutomi, K. et al. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl Acad. Sci. USA 102, 8222–8227 (2005)
Sarin, K. Y. et al. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 436, 1048–1052 (2005)
Lee, J. et al. TERT promotes cellular and organismal survival independently of telomerase activity. Oncogene 27, 3754–3760 (2008)
Tollervey, D. & Kiss, T. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9, 337–342 (1997)
Ridanpaa, M. et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 104, 195–203 (2001)
Moriarty, T. J., Huard, S., Dupuis, S. & Autexier, C. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 22, 1253–1265 (2002)
Lue, N. F. et al. Telomerase can act as a template- and RNA-independent terminal transferase. Proc. Natl Acad. Sci. USA 102, 9778–9783 (2005)
Nakamura, T. M. et al. Telomerase catalytic subunit homologs from fission yeast and human. Science 277, 955–959 (1997)
Mourrain, P. et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542 (2000)
Smardon, A. et al. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans . Curr. Biol. 10, 169–178 (2000)
Lipardi, C., Wei, Q. & Paterson, B. M. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107, 297–307 (2001)
Sijen, T. et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476 (2001)
Makeyev, E. V. & Bamford, D. H. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol. Cell 10, 1417–1427 (2002)
Semler, B. L. & Wimmer, E. Molecular Biology of Picornaviruses 255–267 (American Society for Microbiology, 2002)
Behrens, S. E., Tomei, L. & De Francesco, R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 15, 12–22 (1996)
Sugiyama, T., Cam, H., Verdel, A., Moazed, D. & Grewal, S. I. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl Acad. Sci. USA 102, 152–157 (2005)
Masutomi, K. et al. Telomerase maintains telomere structure in normal human cells. Cell 114, 241–253 (2003)
Ford, L. P. et al. Telomerase can inhibit the recombination-based pathway of telomere maintenance in human cells. J. Biol. Chem. 276, 32198–32203 (2001)
Pascale, E., Cimino Reale, G. & D’Ambrosio, E. Tumor cells fail to trans-induce telomerase in human umbilical vein endothelial cell cultures. Cancer Res. 64, 7702–7705 (2004)
Won, J., Chang, S., Oh, S. & Kim, T. K. Small-molecule-based identification of dynamic assembly of E2F-pocket protein-histone deacetylase complex for telomerase regulation in human cells. Proc. Natl Acad. Sci. USA 101, 11328–11333 (2004)
Almeida, R. & Allshire, R. C. RNA silencing and genome regulation. Trends Cell Biol. 15, 251–258 (2005)
Schwarz, D. S., Hutvagner, G., Haley, B. & Zamore, P. D. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10, 537–548 (2002)
Schwarz, D. S., Tomari, Y. & Zamore, P. D. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr. Biol. 14, 787–791 (2004)
Vagin, V. V. et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 (2006)
Nishikura, K. A short primer on RNAi: RNA-directed RNA polymerase acts as a key catalyst. Cell 107, 415–418 (2001)
Aoki, K., Moriguchi, H., Yoshioka, T., Okawa, K. & Tabara, H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans . EMBO J. 26, 5007–5019 (2007)
Gillis, A. J., Schuller, A. P. & Skordalakes, E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 455, 633–637 (2008)
Salgado, P. S. et al. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 4, e434 (2006)
Tam, O. H. et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453, 534–538 (2008)
Watanabe, T. et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543 (2008)
Babiarz, J. E., Ruby, J. G., Wang, Y., Bartel, D. P. & Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 22, 2773–2785 (2008)
Yang, N. & Kazazian, H. H. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nature Struct. Mol. Biol. 13, 763–771 (2006)
Guggenheim, R., Somech, R., Grunebaum, E., Atkinson, A. & Roifman, C. M. Bone marrow transplantation for cartilage-hair-hypoplasia. Bone Marrow Transplant 38, 751–756 (2006)
Choi, J. et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 4, e10 (2008)
Hahn, W. C. et al. Creation of human tumour cells with defined genetic elements. Nature 400, 464–468 (1999)
Hahn, W. C. et al. Inhibition of telomerase limits the growth of human cancer cells. Nature Med. 5, 1164–1170 (1999)
Kim, N. W. et al. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994)
Yamashita, T. et al. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273, 15479–15486 (1998)
Azuma-Mukai, A. et al. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl Acad. Sci. USA 105, 7964–7969 (2008)
Acknowledgements
We thank T. Sugimura and S. Hirohashi at the National Cancer Center for comments. We also thank H. Siomi, H. Tabara and Y. Tomari for discussions. This work was supported in part by Grant-in-Aid for Young Scientists (A) 19689010 (K.M.) and Grant-in-Aid for Young Scientists (B) 19791141 (Y.M.) from the Ministry of Education, Culture, Sports, Science and Technology, by the Third-Term Comprehensive Control Research for Cancer (K.M.) from the Ministry of Health, Labor, and Welfare, by a Takeda Science Foundation grant (K.M.), the Uehara Memorial Foundation (K.M.), a J&J Focused Funding Award (W.C.H.) and R01 AG23145 from the National Institutes of Health (W.C.H.).
Author Contributions Y.M., M.Y., M.F., N.O., R.P. and V.K. performed experiments. T.L. and Y.H. designed and carried out the bioinformatics analyses of TERT-associated RNAs. Y.M., M.Y., W.C.H. and K.M. designed the experiments and discussed the interpretation of the results. W.C.H. and K.M. cowrote the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-18 with Legends and Supplementary Table 1. (PDF 1604 kb)
Rights and permissions
About this article
Cite this article
Maida, Y., Yasukawa, M., Furuuchi, M. et al. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 461, 230–235 (2009). https://doi.org/10.1038/nature08283
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08283
This article is cited by
-
The regulations of telomerase reverse transcriptase (TERT) in cancer
Cell Death & Disease (2024)
-
M6A RNA methylation-mediated RMRP stability renders proliferation and progression of non-small cell lung cancer through regulating TGFBR1/SMAD2/SMAD3 pathway
Cell Death & Differentiation (2023)
-
The role of alternative splicing in lung cancer
Cancer Chemotherapy and Pharmacology (2023)
-
SMG6 regulates DNA damage and cell survival in Hippo pathway kinase LATS2-inactivated malignant mesothelioma
Cell Death Discovery (2022)
-
Telomerase is required for glomerular renewal in kidneys of adult mice
npj Regenerative Medicine (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.