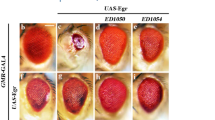
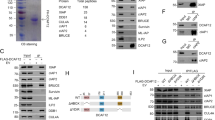
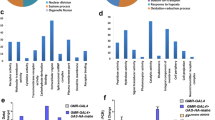
Abstract
Apoptosis is a conserved form of programmed cell death firmly established in the aetiology, pathogenesis and treatment of many human diseases. Central to the core machinery of apoptosis are the caspases and their proximal regulators. Current models for caspase control involve a balance of opposing elements, with variable contributions from positive and negative regulators among different cell types and species1. To advance a comprehensive view of components that support caspase-dependent cell death, we conducted a genome-wide silencing screen in the Drosophila model. Our strategy used a library of double-stranded RNAs together with a chemical antagonist of Inhibitor of apoptosis proteins (IAPs) that simulates the action of native regulators in the Reaper and Smac (also known as Diablo) families2. Here we present a highly validated set of targets that is necessary for death provoked by several stimuli. Among these, Tango7 is identified as a new effector. Cells depleted for this gene resisted apoptosis at a step before the induction of effector caspase activity, and the directed silencing of Tango7 in Drosophila prevented caspase-dependent programmed cell death. Unlike known apoptosis regulators in this model system3, Tango7 activity did not influence stimulus-dependent loss of Drosophila DIAP1 (also known as th and IAP1), but instead regulated levels of the apical caspase Dronc (Nc). Similarly, the human Tango7 counterpart, PCID1 (also known as EIF3M), impinged on caspase 9, revealing a new regulatory axis affecting the apoptosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Salvesen, G. S. & Abrams, J. M. Caspase activation—stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene 23, 2774–2784 (2004)
Verhagen, A. M. & Vaux, D. L. Cell death regulation by the mammalian IAP antagonist Diablo/Smac. Apoptosis 7, 163–166 (2002)
Hay, B. A. & Guo, M. Caspase-dependent cell death in Drosophila . Annu. Rev. Cell Dev. Biol. 22, 623–650 (2006)
Li, L. et al. A small molecule Smac mimic potentiates TRAIL- and TNFα-mediated cell death. Science 305, 1471–1474 (2004)
Shi, Y. Caspase activation, inhibition, and reactivation: a mechanistic view. Protein Sci. 13, 1979–1987 (2004)
Chew, S. K. et al. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila . Dev. Cell 7, 897–907 (2004)
Akdemir, F. et al. Autophagy occurs upstream or parallel to the apoptosome during histolytic cell death. Development 133, 1457–1465 (2006)
Zhang, J. H., Chung, T. D. & Oldenburg, K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4, 67–73 (1999)
Boutros, M. et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303, 832–835 (2004)
Kulkarni, M. M. et al. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nature Methods 3, 833–838 (2006)
Ma, Y., Creanga, A., Lum, L. & Beachy, P. A. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443, 359 (2006)
Zimmermann, K. C., Ricci, J. E., Droin, N. M. & Green, D. R. The role of ARK in stress-induced apoptosis in Drosophila cells. J. Cell Biol. 156, 1077–1087 (2002)
Xu, D. et al. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila . Development 132, 2125–2134 (2005)
Muro, I. et al. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development 133, 3305–3315 (2006)
Link, N. et al. A collective form of cell death requires homeodomain interacting protein kinase. J. Cell Biol. 178, 567–574 (2007)
Rodriguez, A. et al. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nature Cell Biol. 1, 272–279 (1999)
Muro, I., Hay, B. A. & Clem, R. J. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J. Biol. Chem. 277, 49644–49650 (2002)
Weinmann, L. et al. A novel p53 rescue compound induces p53-dependent growth arrest and sensitises glioma cells to Apo2L/TRAIL-induced apoptosis. Cell Death Differ. 15, 718–729 (2008)
Matapurkar, A. & Lazebnik, Y. Requirement of cytochrome c for apoptosis in human cells. Cell Death Differ. 13, 2062–2067 (2006)
Kroemer, G. & Martin, S. J. Caspase-independent cell death. Nature Med. 11, 725–730 (2005)
Colell, A. et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 129, 983–997 (2007)
Bard, F. et al. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439, 604–607 (2006)
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008)
Perez, A. et al. A new class of receptor for herpes simplex virus has heptad repeat motifs that are common to membrane fusion proteins. J. Virol. 79, 7419–7430 (2005)
Murray, T. V. et al. A non-apoptotic role for caspase-9 in muscle differentiation. J. Cell Sci. 121, 3786–3793 (2008)
Malo, N. et al. Statistical practice in high-throughput screening data analysis. Nature Biotechnol. 24, 167–175 (2006)
Clemens, J. C. et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl Acad. Sci. USA 97, 6499–6503 (2000)
Mendes, C. S. et al. Cytochrome c-d regulates developmental apoptosis in the Drosophila retina. EMBO Rep. 7, 933–939 (2006)
Ditzel, M. et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nature Cell Biol. 5, 467–473 (2003)
Acknowledgements
We thank X. Tu, L. Durham, L. Lum, P. Beachy, M. Roth, X. Wang, P. Harran, Y. Lazebnik, D. Dorris, the University of Texas Southwestern Medical Center (UTSW) High Throughput Screening laboratory and the Live Cell Imaging facility for technical and material support; Bloomington Stock Center, K. Basler, National Institute of Genetics (NIG) stock center (Japan) for fly lines; K. White, B. Hay and the Hybridoma Bank for antibodies; V. Malhotra for the pMT-Tango7 plasmid; J. Seeman and the Wang laboratory for discussions; and M. Allen for administrative support. This work was supported by grants from the National Institute of General Medical Sciences, the National Institute on Alcohol Abuse and Alcoholism and the UTSW High Impact/High Risk Grant Program. N.L. is supported by a National Research Service Award.
Author Contributions S.K.C., N.L., P.C., K.A.G. and K.P. planned and performed experiments and analysed results. All authors discussed results. J.M.A. planned experiments, analysed results and wrote the paper together with S.K.C., aided by contributions from N.L. and P.C.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 with Legend and Supplementary Tables 1-3. (PDF 398 kb)
Supplementary Table
This file contains Supplementary Table 4 which shows dsRNA Library Sequences. (XLS 9605 kb)
Supplementary Movie 1
This movie shows S2R+ cells treated with Smac-mimetic compound to induce apoptosis. Time lapse covers period of 12 hrs. (MOV 4144 kb)
Supplementary Movie 2
This movie shows S2R+ cells treated with UV irradiation and control AmpR dsRNA. Time lapse covers period of 22 hrs. (MOV 7758 kb)
Supplementary Movie 3
This movie shows S2R+ cells treated with UV irradiation and Dronc dsRNA. Time lapse covers period of 22 hrs. (MOV 5859 kb)
Supplementary Movie 4
This movie shows S2R+ cells treated with UV irradiation and Tango7 dsRNA. Time lapse covers period of 22 hrs. (MOV 9419 kb)
Rights and permissions
About this article
Cite this article
Chew, S., Chen, P., Link, N. et al. Genome-wide silencing in Drosophila captures conserved apoptotic effectors. Nature 460, 123–127 (2009). https://doi.org/10.1038/nature08087
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08087
This article is cited by
-
miR-277 targets the proapoptotic gene-hid to ameliorate Aβ42-mediated neurodegeneration in Alzheimer’s model
Cell Death & Disease (2024)
-
Caspase-dependent non-apoptotic processes in development
Cell Death & Differentiation (2017)
-
Killers creating new life: caspases drive apoptosis-induced proliferation in tissue repair and disease
Cell Death & Differentiation (2017)
-
Tango7 regulates cortical activity of caspases during reaper-triggered changes in tissue elasticity
Nature Communications (2017)
-
The de-ubiquitylating enzyme DUBA is essential for spermatogenesis in Drosophila
Cell Death & Differentiation (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.