Abstract
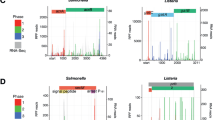
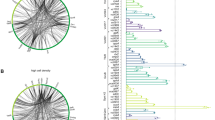
The bacterium Listeria monocytogenes is ubiquitous in the environment and can lead to severe food-borne infections. It has recently emerged as a multifaceted model in pathogenesis. However, how this bacterium switches from a saprophyte to a pathogen is largely unknown. Here, using tiling arrays and RNAs from wild-type and mutant bacteria grown in vitro, ex vivo and in vivo, we have analysed the transcription of its entire genome. We provide the complete Listeria operon map and have uncovered far more diverse types of RNAs than expected: in addition to 50 small RNAs (<500 nucleotides), at least two of which are involved in virulence in mice, we have identified antisense RNAs covering several open-reading frames and long overlapping 5′ and 3′ untranslated regions. We discovered that riboswitches can act as terminators for upstream genes. When Listeria reaches the host intestinal lumen, an extensive transcriptional reshaping occurs with a SigB-mediated activation of virulence genes. In contrast, in the blood, PrfA controls transcription of virulence genes. Remarkably, several non-coding RNAs absent in the non-pathogenic species Listeria innocua exhibit the same expression patterns as the virulence genes. Together, our data unravel successive and coordinated global transcriptional changes during infection and point to previously unknown regulatory mechanisms in bacteria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
ArrayExpress
Data deposits
Raw data are available from ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-MEXP-2138 for gene expression sub-array analysis and E-MEXP-2142 for tiling sub-array analysis.
References
Cossart, P. & Toledo-Arana, A. Listeria monocytogenes, a unique model in infection biology: an overview. Microbes Infect. 10, 1041–1050 (2008)
Lecuit, M. Understanding how Listeria monocytogenes targets and crosses host barriers. Clin. Microbiol. Infect. 11, 430–436 (2005)
Hamon, M., Bierne, H. & Cossart, P. Listeria monocytogenes: a multifaceted model. Nature Rev. Microbiol. 4, 423–434 (2006)
Glaser, P. et al. Comparative genomics of Listeria species. Science 294, 849–852 (2001)
Dussurget, O., Pizarro-Cerda, J. & Cossart, P. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58, 587–610 (2004)
Leimeister-Wachter, M. et al. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes . Proc. Natl Acad. Sci. USA 87, 8336–8340 (1990)
Mengaud, J. et al. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5, 2273–2283 (1991)
Johansson, J. et al. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes . Cell 110, 551–561 (2002)
Garner, M. R., Njaa, B. L., Wiedmann, M. & Boor, K. J. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect. Immun. 74, 876–886 (2006)
Gahan, C. G. & Hill, C. Gastrointestinal phase of Listeria monocytogenes infection. J. Appl. Microbiol. 98, 1345–1353 (2005)
Mandin, P. et al. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 57, 1367–1380 (2005)
Barry, T., Kelly, M., Glynn, B. & Peden, J. Molecular cloning and phylogenetic analysis of the small cytoplasmic RNA from Listeria monocytogenes . FEMS Microbiol. Lett. 173, 47–53 (1999)
Christiansen, J. K. et al. Identification of small Hfq-binding RNAs in Listeria monocytogenes . RNA 12, 1383–1396 (2006)
Mandin, P. et al. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 35, 962–974 (2007)
Nielsen, J. S. et al. Identification of a sigma B-dependent small noncoding RNA in Listeria monocytogenes . J. Bacteriol. 190, 6264–6270 (2008)
Toledo-Arana, A., Repoila, F. & Cossart, P. Small noncoding RNAs controlling pathogenesis. Curr. Opin. Microbiol. 10, 182–188 (2007)
Christiansen, J. K. et al. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 186, 3355–3362 (2004)
Yazaki, J., Gregory, B. D. & Ecker, J. R. Mapping the genome landscape using tiling array technology. Curr. Opin. Plant Biol. 10, 534–542 (2007)
Gregory, B. D., Yazaki, J. & Ecker, J. R. Utilizing tiling microarrays for whole-genome analysis in plants. Plant J. 53, 636–644 (2008)
David, L. et al. A high-resolution map of transcription in the yeast genome. Proc. Natl Acad. Sci. USA 103, 5320–5325 (2006)
Selinger, D. W. et al. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nature Biotechnol. 18, 1262–1268 (2000)
McGrath, P. T. et al. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nature Biotechnol. 25, 584–592 (2007)
Landt, S. G. et al. Small non-coding RNAs in Caulobacter crescentus . Mol. Microbiol. 68, 600–614 (2008)
Disson, O. et al. Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature 455, 1114–1118 (2008)
Winkler, W. C. Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Curr. Opin. Chem. Biol. 9, 594–602 (2005)
Brantl, S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 10, 102–109 (2007)
Coppins, R. L., Hall, K. B. & Groisman, E. A. The intricate world of riboswitches. Curr. Opin. Microbiol. 10, 176–181 (2007)
Griffiths-Jones, S. et al. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33 (Database issue). D121–D124 (2005)
Loh, E., Gripenland, J. & Johansson, J. Control of Listeria monocytogenes virulence by 5′-untranslated RNA. Trends Microbiol. 14, 294–298 (2006)
Grundling, A., Burrack, L. S., Bouwer, H. G. & Higgins, D. E. Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc. Natl Acad. Sci. USA 101, 12318–12323 (2004)
Raengpradub, S., Wiedmann, M. & Boor, K. J. Comparative analysis of the sigma B-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl. Environ. Microbiol. 74, 158–171 (2008)
Kazmierczak, M. J., Mithoe, S. C., Boor, K. J. & Wiedmann, M. Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185, 5722–5734 (2003)
Hain, T. et al. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e sigma B regulon. BMC Microbiol. 8, 20 (2008)
Dussurget, O. et al. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45, 1095–1106 (2002)
Chakraborty, T., Hain, T. & Domann, E. Genome organization and the evolution of the virulence gene locus in Listeria species. Int. J. Med. Microbiol. 290, 167–174 (2000)
Scortti, M. et al. The PrfA virulence regulon. Microbes Infect. 9, 1196–1207 (2007)
Chico-Calero, I. et al. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria . Proc. Natl Acad. Sci. USA 99, 431–436 (2002)
Acknowledgements
We are grateful to E. Charpentier and the members of her group for providing the RACE protocol. We thank L. Frangeul for helping with L. monocytogenes annotation files. J.J. is supported by the Swedish Research Council grants K2008-58X-15144-05-3 and 621-2006-4450 and EU (BacRNA 2005 Contract N° 018618). Work in the laboratory of P.C. received financial support from Institut Pasteur (GPH 9), Inserm, INRA, EU (BacRNA 2005-018618), ANR (ANR-05-MIIM-026-01) and ERC (Advanced Grant 233348). A.T.-A. was an EMBO long-term fellow. P.C. is an international research scholar of the Howard Hughes Medical Institute.
Author Contributions P.C. planned the project. A.T.-A., O.D., M.L. and P.C. designed the research. A.T.-A., O.D., G.N., N.S., H.G.-R., D.B., E.L., J.G., T.T., K.V., M.-A.N., G.S. and M.L. performed the experiments. A.T.-A., O.D., M.B., M.V., B.R., J.-Y.C., M.L., J.J. and P.C. analysed the experiments. A.T.-A., O.D. and P.C. wrote the paper and co-authors commented on it.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Materials and Methods, Supplementary References and Supplementary Tables S1-S5. (PDF 765 kb)
Supplementary Table
This file contains Supplementary Table 6. (XLS 485 kb)
Supplementary Figures
This file contains Supplementary Figures S1-S16 with Legends. (PDF 6908 kb)
Supplementary Figures
This file contains Supplementary Figures S17-S32 with Legends. (PDF 18172 kb)
Supplementary Figures
This file contains Supplementary Figures S33-S40 with Legends. (PDF 14585 kb)
Supplementary Figures
This file contains Supplementary Figures S41-48 with Legends. (PDF 14812 kb)
Rights and permissions
About this article
Cite this article
Toledo-Arana, A., Dussurget, O., Nikitas, G. et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956 (2009). https://doi.org/10.1038/nature08080
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08080
This article is cited by
-
Harmonization of supervised machine learning practices for efficient source attribution of Listeria monocytogenes based on genomic data
BMC Genomics (2023)
-
Design and validation of a dual-fluorescence reporter system to monitor bacterial gene expression in the gut environment
Applied Microbiology and Biotechnology (2023)
-
Immunopeptidomics-based design of mRNA vaccine formulations against Listeria monocytogenes
Nature Communications (2022)
-
Transcriptome architecture and regulation at environmental transitions in flavobacteria: the case of an important fish pathogen
ISME Communications (2021)
-
Metagenomic and metatranscriptomic profiling of Lactobacillus casei Zhang in the human gut
npj Biofilms and Microbiomes (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.