Abstract
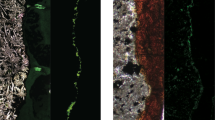
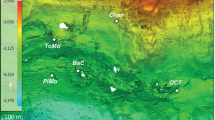
Oceanic lithosphere exposed at the sea floor undergoes seawater–rock alteration reactions involving the oxidation and hydration of glassy basalt. Basalt alteration reactions are theoretically capable of supplying sufficient energy for chemolithoautotrophic growth1. Such reactions have been shown to generate microbial biomass in the laboratory2, but field-based support for the existence of microbes that are supported by basalt alteration is lacking. Here, using quantitative polymerase chain reaction, in situ hybridization and microscopy, we demonstrate that prokaryotic cell abundances on seafloor-exposed basalts are 3–4 orders of magnitude greater than in overlying deep sea water. Phylogenetic analyses of basaltic lavas from the East Pacific Rise (9° N) and around Hawaii reveal that the basalt-hosted biosphere harbours high bacterial community richness and that community membership is shared between these sites. We hypothesize that alteration reactions fuel chemolithoautotrophic microorganisms, which constitute a trophic base of the basalt habitat, with important implications for deep-sea carbon cycling and chemical exchange between basalt and sea water.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bach, W. & Edwards, K. J. Iron and sulfide oxidation within the basaltic ocean crust: Implications for chemolithoautotrophic microbial biomass production. Geochim. Cosmochim. Acta 67, 3871–3887 (2003)
Edwards, K. J., Rogers, D. R., Wirsen, C. O. & McCollom, T. M. Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic α- and γ-Proteobacteria from the deep sea. Appl. Environ. Microbiol. 69, 2906–2913 (2003)
Karner, M. B., DeLong, E. F. & Karl, D. M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510 (2001)
Cowen, J. P. et al. Fluids from aging ocean crust that support microbial life. Science 299, 120–123 (2003)
Kormas, K. A., Tivey, M. K., Von Damm, K. & Teske, A. Bacterial and archaeal phylotypes associated with distinct mineralogical layers of a white smoker spire from a deep-sea hydrothermal vent site (9° N, East Pacific Rise). Environ. Microbiol. 8, 909–920 (2006)
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004)
Reysenbach, A.-L., Longnecker, K. & Kirshtein, J. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a mid-atlantic ridge hydrothermal vent. Appl. Environ. Microbiol. 66, 3798–3806 (2000)
Kormas, K. A., Smith, D. C., Edgcomb, V. & Teske, A. Molecular analysis of deep subsurface microbial communities in Nankai Trough sediments (ODP Leg 190, Site 1176). FEMS Microbiol. Ecol. 45, 115–125 (2003)
Tringe, S. G. et al. Comparative metagenomics of microbial communities. Science 308, 554–557 (2005)
Ley, R. E. et al. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 72, 3685–3695 (2006)
Hong, S.-H., Bunge, J., Jeon, S.-O. & Epstein, S. S. Predicting microbial species richness. Proc. Natl Acad. Sci. USA 103, 117–122 (2006)
Hughes, J., Hellmann, J., Ricketts, T. & Bohannan, B. Counting the uncountable: statistical approaches to estimating microbial diversity. Environ. Microbiol. 67, 4399–4406 (2001)
Hughes, J. B. & Hellmann, J. J. The application of rarefaction techniques to molecular inventories of microbial diversity. Methods Enzymol. 397, 292–308 (2005)
Huber, J. A., Butterfield, D. A. & Baross, J. A. Bacterial diversity in a subseafloor habitat following a deep-sea volcanic eruption. FEMS Microbiol. Ecol. 43, 393–409 (2003)
Huber, J. A., Johnson, H. P., Butterfield, D. A. & Baross, J. A. Microbial life in ridge flank crustal fluids. Environ. Microbiol. 8, 88–99 (2006)
Huber, J. A. et al. Microbial population structure in the deep marine biosphere. Science 318, 97–100 (2007)
Edwards, K. J., Bach, W. & McCollom, T. M. Geomicrobiology in oceanography: mineral–microbe interactions at and below the seafloor. Trends Microbiol. 13, 449–459 (2005)
Lang, S. Q., Butterfield, D. A., Lilley, M. D., Johnson, H. P. & Hedges, J. I. Dissolved organic carbon in ridge-axis and ridge-flank hydrothermal systems. Geochim. Cosmochim. Acta 70, 3830–3842 (2006)
Barber, R. T. Dissolved organic carbon from deep waters resists microbial oxidation. Nature 220, 274–275 (1968)
Whitman, W. B., Coleman, D. C. & Wiebe, W. J. Prokaryotes: The unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583 (1998)
Biddle, J. F. et al. Heterotrophic archaea dominate sedimentary subsurface ecosystems of Peru. Proc. Natl Acad. Sci. USA 103, 3846–3851 (2006)
Rogers, D. R., Santelli, C. M. & Edwards, K. J. Geomicrobiology of deep-sea deposits: estimating community diversity from low-temperature seafloor rocks and minerals. Geobiology 1, 109–117 (2003)
Cole, J. R. et al. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31, 442–443 (2003)
Schloss, P. D. & Handelsman, J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71, 1501–1506 (2005)
Schloss, P. D. & Handelsman, J. Introducing SONS, a tool for OTU-based comparisons of membership and structure between microbial communities. Appl. Environ. Microbiol. 72, 6773–6779 (2006)
Daims, H., Bruhl, A., Amann, R., Schleifer, K.-H. & Wagner, M. Probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22, 438–448 (1999)
Raskin, L., Stromley, J., Rittmann, B. E. & Stahl, D. A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl. Environ. Microbiol. 60, 1232–1240 (1994)
Nadkarni, M., Martin, F. E., Jacques, N. A. & Hunter, N. Determination of bacterial load by real-time PCR using a broad range (universal) probe and primer set. Microbiology 148, 257–266 (2002)
Takai, K. & Horikoshi, K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66, 5066–5072 (2000)
Klappenbach, J. A., Saxman, P. R., Cole, J. R. & Schmidt, T. M. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 29, 181–184 (2001)
Webster, G., Newberry, C. J., Fry, J. C. & Weightman, A. J. Assessment of bacterial community structure in the deep sub-seafloor biosphere by 16S rDNA-based techniques: a cautionary tale. J. Microbiol. Methods 55, 155–164 (2003)
Huber, T., Faulkner, G. & Hugenholtz, P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20, 2317–2319 (2004)
Ashelford, K. E., Chuzhanova, N. A., Fry, J. C., Jones, A. J. & Weightman, A. J. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72, 5734–5741 (2006)
Altschul, S. F. et al. Gapped BLAST and PSI-blast: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997)
Davis, R. E. & Moyer, C. L. Extreme spatial variability in microbial mat communities from submarine hydrothermal vents located at multiple volcanoes along the Mariana Island. Eos. Trans. AGU 86, Fall Meet. Suppl Abstract V51C–1509 (2005)
Pernthaler, A., Pernthaler, J. & Amann, R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68, 3094–3101 (2002)
Sekar, R. et al. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69, 2928–2935 (2003)
Acknowledgements
The authors thank D. Fornari, M. Tivey and H. Schouten for allowing C.M.S. and W.B. to participate in their research cruise AT11-7 to collect samples at the EPR, D. Rogers for samples collected on the research cruise KOK 02-24 at Hawaii, L. Kerr for instruction and guidance on the SEM and confocal microscope, P. Schloss for support with the DOTUR and SONS programs, S. Simmons for support and instruction on qPCR, and E. Leadbetter for advice on the manuscript. This research was supported by a Ridge2K grant awarded to K.J.E. and W.B., in part by a NAI CAN awarded to M.L.S. and K.J.E., and also in part by a Project Development Award from WWU’s Office of Research and Sponsored Programs to C.L.M.
Author Contributions Phylogenetic analyses were performed by C.M.S. for EPR samples, and B.N.O. and E.B. for Hawaii samples; microscopy was done by C.M.S. Biomass calculations were done by W.B. C.L.M. helped C.M.S. with qPCR analyses. M.L.S. supported Hawaii studies and the pipeline for phylogenetic studies. H.S. provided ship access for Hawaii studies. K.J.E. and W.B. developed and guided this project, and K.J.E., C.M.S. and B.N.O. developed and wrote the paper with input from co-authors.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
The file contains Supplementary Discussion referred to in the manuscript, Supplementary Tables S1-S3, Supplementary Figures S1-S6 with Legends, and additional references. (PDF 749 kb)
Rights and permissions
About this article
Cite this article
Santelli, C., Orcutt, B., Banning, E. et al. Abundance and diversity of microbial life in ocean crust. Nature 453, 653–656 (2008). https://doi.org/10.1038/nature06899
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06899
This article is cited by
-
Chemolithotrophic biosynthesis of organic carbon associated with volcanic ash in the Mariana Trough, Pacific Ocean
Communications Earth & Environment (2023)
-
Insights into Bacterial Community Structure and Metabolic Diversity of Mercury-Contaminated Sediments from Hyeongsan River, Pohang, South Korea
Current Microbiology (2022)
-
Bacterial communities in temperate and polar coastal sands are seasonally stable
ISME Communications (2021)
-
Modern precipitation of hydrogenetic ferromanganese minerals during on-site 15-year exposure tests
Scientific Reports (2020)
-
Iron-oxidizer hotspots formed by intermittent oxic–anoxic fluid mixing in fractured rocks
Nature Geoscience (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.