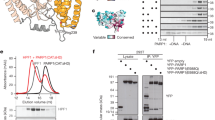
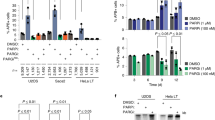
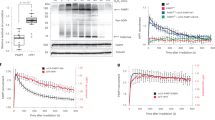
Abstract
Post-translational modification (PTM) of proteins plays an important part in mediating protein interactions and/or the recruitment of specific protein targets1,2. PTM can be mediated by the addition of functional groups (for example, acetylation or phosphorylation), peptides (for example, ubiquitylation or sumoylation), or nucleotides (for example, poly(ADP-ribosyl)ation). Poly(ADP-ribosyl)ation often involves the addition of long chains of ADP-ribose units, linked by glycosidic ribose–ribose bonds3, and is critical for a wide range of processes, including DNA repair, regulation of chromosome structure, transcriptional regulation, mitosis and apoptosis4. Here we identify a novel poly(ADP-ribose)-binding zinc finger (PBZ) motif in a number of eukaryotic proteins involved in the DNA damage response and checkpoint regulation. The PBZ motif is also required for post-translational poly(ADP-ribosyl)ation. We demonstrate interaction of poly(ADP-ribose) with this motif in two representative human proteins, APLF (aprataxin PNK-like factor) and CHFR (checkpoint protein with FHA and RING domains), and show that the actions of CHFR in the antephase checkpoint are abrogated by mutations in PBZ or by inhibition of poly(ADP-ribose) synthesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Walsh, C. T. Postranslational Modification of Proteins: Expanding Nature's Inventory (Roberts and Co., Greenwood Village, Colorado, 2006)
Seet, B. T. et al. Reading protein modifications with interaction domains. Nature Rev. Mol. Cell Biol. 7, 473–483 (2006)
D'Amours, D. et al. Poly(ADP-ribosyl)ation reaction in the regulation of nuclear functions. J. Biochem. 342, 249–268 (1999)
Kim, M. Y., Zhang, T. & Kraus, W. L. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev. 19, 1951–1967 (2005)
Hassa, P. O. et al. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev. 70, 789–829 (2006)
Pleschke, J. M. et al. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275, 40974–40980 (2000)
Gagne, J. P. et al. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. Biochem. J. 371, 331–340 (2003)
Karras, G. I. et al. The macro domain is an ADP-ribose binding module. EMBO J. 24, 1911–1920 (2005)
Matsusaka, T. & Pines, J. CHFR acts with the p38 stress kinases to block entry to mitosis in mammalian cells. J. Cell Biol. 166, 507–516 (2004)
Scolnick, D. M. & Halazonetis, T. D. CHFR defines a mitotic stress checkpoint that delays entry into metaphase. Nature 406, 430–435 (2000)
Kang, D. et al. The checkpoint protein CHFR is a ligase that ubiquitinates Plk1 and inhibits Cdc2 at the G2 to M transition. J. Cell Biol. 156, 249–259 (2002)
Yu, X. et al. CHFR is required for tumour suppression and Aurora A regulation. Nature Genet. 37, 401–406 (2005)
Iles, N. et al. APLF (C2orf13) is a novel protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 27, 3793–3803 (2007)
Kanno, S.-I. et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 26, 2094–2103 (2007)
Bekker-Jensen, S. et al. Human XIP1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J. Biol. Chem. 282, 19638–19643 (2007)
Masson, M. et al. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA-damage. Mol. Cell. Biol. 18, 3563–3571 (1998)
Rieder, C. L. & Cole, R. Entry into mitosis in vertebrate somatic cells is guarded by a chromosome damage checkpoint that reverses the cell cycle when triggered during early but not late prophase. J. Cell Biol. 142, 1013–1022 (1998)
Rieder, C. L. & Cole, R. Microscopy-induced radiation damage, microtubules, and progression through the terminal stage of G2 (prophase) in vertebrate somatic cells. Cold Spring Harb. Symp. Quant. Biol. 65, 369–376 (2000)
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005)
Miller, J., McLachlan, A. D. & Klug, A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4, 1609–1614 (1985)
Gamsjaeger, R. et al. Sticky fingers: zinc fingers as protein recognition motifs. Trends Biochem. Sci. 32, 63–70 (2007)
Minaga, T. & Kun, E. Probable helical conformation of poly(ADP-ribose). The effect of cations on spectral properties. J. Biol. Chem. 258, 5726–5730 (1983)
Leppard, J. B. et al. Physical and functional interaction between DNA ligase IIIα and poly(ADP-ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell. Biol. 23, 5919–5927 (2003)
Galande, S. & Kohwi-Shigematsu, T. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 274, 20521–20528 (1999)
Burkle, A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ . FEBS J. 272, 4576–4589 (2005)
Bothos, J. et al. The CHFR checkpoint protein functions with Ubc13-Mms2 to form Lys63-linked polyubiquitin chains. Oncogene 22, 7101–7107 (2003)
Acknowledgements
We thank T. Lindahl (LRI, CRUK) for XRCC1, G. Smith for the PARP inhibitor KU-0058948, and J. Gannon for assistance with the Biacore. This work was supported by Cancer Research UK, the EU DNA Repair Consortium and the Louis-Jeantet Foundation. I.A. and D.A. are supported by EMBO fellowships.
Author Contributions I.A. and D.A. discovered the PBZ motif and performed most of the experiments. A.J.C. carried out supporting analyses. T.M. and J.P. defined the role of PBZ in the antephase checkpoint. S.J.B. and S.C.W. are joint senior authors who managed the project and helped write the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Figures 1-2 and Supplementary Table 1. (PDF 3838 kb)
Supplementary Video 1
This file contains Supplementary Video 1. (MOV 11843 kb)
Supplementary Video 2
This file contains Supplementary Video 2. (MOV 16953 kb)
Supplementary Video 3
This file contains Supplementary Video 3. (MOV 4392 kb)
Rights and permissions
About this article
Cite this article
Ahel, I., Ahel, D., Matsusaka, T. et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451, 81–85 (2008). https://doi.org/10.1038/nature06420
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06420
This article is cited by
-
PARP14 correlates with GBM proliferation and poor prognosis by elevating expression of SAMD/SAMD9L
Irish Journal of Medical Science (1971 -) (2024)
-
Poly(ADP-ribose)-binding protein RCD1 is a plant PARylation reader regulated by Photoregulatory Protein Kinases
Communications Biology (2023)
-
Multiple E3 ligases control tankyrase stability and function
Nature Communications (2023)
-
Ubiquitin ligase CHFR mediated degradation of VE-cadherin through ubiquitylation disrupts endothelial adherens junctions
Nature Communications (2023)
-
Horizontal acquisition of a DNA ligase improves DNA damage tolerance in eukaryotes
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.