Abstract
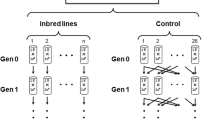
Spontaneous mutations are the source of genetic variation required for evolutionary change, and are therefore important for many aspects of evolutionary biology. For example, the divergence between taxa at neutrally evolving sites in the genome is proportional to the per nucleotide mutation rate, u (ref. 1), and this can be used to date speciation events by assuming a molecular clock. The overall rate of occurrence of deleterious mutations in the genome each generation (U) appears in theories of nucleotide divergence and polymorphism2, the evolution of sex and recombination3, and the evolutionary consequences of inbreeding2. However, estimates of U based on changes in allozymes4 or DNA sequences5 and fitness traits are discordant6,7,8. Here we directly estimate u in Drosophila melanogaster by scanning 20 million bases of DNA from three sets of mutation accumulation lines by using denaturing high-performance liquid chromatography9. From 37 mutation events that we detected, we obtained a mean estimate for u of 8.4 × 10-9 per generation. Moreover, we detected significant heterogeneity in u among the three mutation-accumulation-line genotypes. By multiplying u by an estimate of the fraction of mutations that are deleterious in natural populations of Drosophila10, we estimate that U is 1.2 per diploid genome. This high rate suggests that selection against deleterious mutations may have a key role in explaining patterns of genetic variation in the genome, and help to maintain recombination and sexual reproduction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Kimura, M. The Neutral Theory of Molecular Evolution (Cambridge University Press, Cambridge, 1983)
Charlesworth, B. & Charlesworth, D. Some evolutionary consequences of deleterious mutations. Genetica 102–103, 3–19 (1998)
Kondrashov, A. S. Deleterious mutations and the evolution of sexual reproduction. Nature 336, 435–440 (1988)
Mukai, T. & Cockerham, C. C. Spontaneous mutation rates at enzyme loci in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 74, 2514–2517 (1977)
Denver, D. R., Morris, K., Lynch, M. & Thomas, W. K. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430, 679–682 (2004)
Crow, J. F. & Simmons, M. J. in The Genetics and Biology of Drosophila Vol. 3C (eds Ashburner, M., Carson, H. L. & Thompson, J. N.) 1–35 (Academic, London, 1983)
Keightley, P. D. & Eyre-Walker, A. Terumi Mukai and the riddle of deleterious mutation rates. Genetics 153, 515–523 (1999)
Lynch, M. et al. Perspective: Spontaneous deleterious mutation. Evolution 53, 645–663 (1999)
Oefner, P. J. & Huber, C. G. A decade of high-resolution liquid chromatography of nucleic acids on styrene divinylbenzene copolymers. J. Chromatogr. B 782, 27–55 (2002)
Halligan, D. L. & Keightley, P. D. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 16, 875–884 (2006)
Keightley, P. D. & Otto, S. P. Interference among deleterious mutations favours sex and recombination in finite populations. Nature 443, 89–92 (2006)
Presgraves, D. C. Recombination enhances protein adaptation in Drosophila melanogaster. Curr. Biol. 15, 1651–1656 (2005)
Nachman, M. W. Single nucleotide polymorphisms and recombination rate in humans. Trends Genet. 17, 481–485 (2001)
Kondrashov, A. S. & Crow, J. F. A molecular approach to estimating the human deleterious mutation rate. Hum. Mutat. 2, 229–234 (1993)
Houle, D. & Nuzhdin, S. V. Mutation accumulation and the effect of copia insertions in Drosophila melanogaster. Genet. Res. 83, 7–18 (2004)
Fernandez, J. & López-Fanjul, C. Spontaneous mutational variances and covariances for fitness-related traits in Drosophila melanogaster. Genetics 143, 829–837 (1996)
Maside, X., Bartolome, C., Assimacopoulos, S. & Charlesworth, B. Rates of movement and distribution of transposable elements in Drosophila melanogaster: in situ hybridization vs Southern blotting data. Genet. Res. 78, 121–136 (2001)
Dobson-Stone, C. et al. Comparison of fluorescent single-strand conformation polymorphism analysis and denaturing high-performance liquid chromatography for detection of EXT1 and EXT2 mutations in hereditary multiple exostoses. Eur. J. Hum. Genet. 8, 24–32 (2000)
O’Donovan, M. C. et al. Blind analysis of denaturing high-performance liquid chromatography as a tool for mutation detection. Genomics 52, 44–49 (1998)
Moriyama, E. N. & Powell, J. R. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13, 261–277 (1996)
Petrov, D. A. DNA loss and evolution of genome size in Drosophila. Genetica 115, 81–91 (2002)
Woodruff, R. C., Thompson, J. N., Seeger, M. A. & Spivey, W. E. Variation in spontaneous mutation and repair in natural population lines of Drosophila melanogaster. Heredity 53, 223–234 (1984)
Baer, C. F. et al. Comparative evolutionary genetics of spontaneous mutations affecting fitness in rhabditid nematodes. Proc. Natl Acad. Sci. USA 102, 5785–5790 (2005)
Tamura, K., Subramanian, S. & Kumar, S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol. Biol. Evol. 21, 36–44 (2004)
Andolfatto, P. Adaptive evolution of non-coding DNA in Drosophila. Nature 437, 1149–1152 (2005)
Fry, J. D. On the rate and linearity of viability declines in Drosophila mutation-accumulation experiments: Genomic mutation rates and synergistic epistasis revisited. Genetics 166, 797–806 (2004)
Loewe, L. & Charlesworth, B. Inferring the distribution of mutational effects on fitness in Drosophila. Biol. Lett. 2, 426–430 (2006)
Charlesworth, B. Mutation selection balance and the evolutionary advantage of sex and recombination. Genet. Res. 55, 199–221 (1990)
Salathé, M., Salathé, R., Schmid-Hempel, P. & Bonhoeffer, S. Mutation accumulation in space and the maintenance of sexual reproduction. Ecol. Lett. 9, 941–946 (2006)
Ravnik-Glavač, M., Atkinson, A., Glavač, D. & Dean, M. DHPLC screening of cystic fibrosis gene mutations. Hum. Mutat. 19, 374–383 (2002)
Acknowledgements
We thank D. Houle and C. López-Fanjul for providing samples of MA lines, P. Andolfatto for suggesting the use of PCR errors as positive controls, F. Oliver for help with DNA sequencing, and D. Charlesworth, J. Crow, J. Drake, A. Eyre-Walker, C. Haag, D. Houle and M. Lynch for comments on the manuscript. We are grateful to the Wellcome Trust for funding by a Functional Genomics Development Initiative grant.
Author Contributions S.M., C.H.-L. and M.D. performed the DHPLC analysis. M.D. cloned and sequenced putative variants. X.M. cloned and sequenced positive controls. D.L.H. analysed selective constraints on indel mutations. B.C. advised on Drosophila genetics and interpreting the data. P.D.K. conceived and designed the project. C.H.-L. and P.D.K. analysed the data and wrote the paper. All authors revised the draft manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-4 and Supplementary Figures 1-6. (PDF 2440 kb)
Rights and permissions
About this article
Cite this article
Haag-Liautard, C., Dorris, M., Maside, X. et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445, 82–85 (2007). https://doi.org/10.1038/nature05388
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05388
This article is cited by
-
The origin and structural evolution of de novo genes in Drosophila
Nature Communications (2024)
-
A transposon-based genetic marker for conspecific identity within the Bactrocera dorsalis species complex
Scientific Reports (2024)
-
Selection in a growing colony biases results of mutation accumulation experiments
Scientific Reports (2022)
-
Sexual selection for males with beneficial mutations
Scientific Reports (2022)
-
Is there still evolution in the human population?
Biologia Futura (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.