Abstract
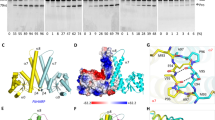
Most eukaryotic messenger RNA precursors (pre-mRNAs) undergo extensive maturational processing, including cleavage and polyadenylation at the 3′-end1,2,3,4,5,6,7,8. Despite the characterization of many proteins that are required for the cleavage reaction, the identity of the endonuclease is not known4,9,10. Recent analyses indicated that the 73-kDa subunit of cleavage and polyadenylation specificity factor (CPSF-73) might be the endonuclease for this and related reactions10,11,12,13,14,15, although no direct data confirmed this. Here we report the crystal structures of human CPSF-73 at 2.1 Å resolution, complexed with zinc ions and a sulphate that might mimic the phosphate group of the substrate, and the related yeast protein CPSF-100 (Ydh1) at 2.5 Å resolution. Both CPSF-73 and CPSF-100 contain two domains, a metallo-β-lactamase domain and a novel β-CASP (named for metallo-β-lactamase, CPSF, Artemis, Snm1, Pso2) domain12. The active site of CPSF-73, with two zinc ions, is located at the interface of the two domains. Purified recombinant CPSF-73 possesses RNA endonuclease activity, and mutations that disrupt zinc binding in the active site abolish this activity. Our studies provide the first direct experimental evidence that CPSF-73 is the pre-mRNA 3′-end-processing endonuclease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Colgan, D. F. & Manley, J. L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 11, 2755–2766 (1997)
Wahle, E. & Ruegsegger, U. 3'-end processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 23, 277–295 (1999)
Zhao, J., Hyman, L. & Moore, C. L. Formation of mRNA 3' ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63, 405–445 (1999)
Proudfoot, N. J. New perspectives on connecting messenger RNA 3' end formation to transcription. Curr. Opin. Cell Biol. 16, 272–278 (2004)
Zorio, D. A. R. & Bentley, D. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 296, 91–97 (2004)
Wilusz, C. J., Wormington, M. & Peltz, S. W. The cap-to-tail guide to mRNA turnover. Nature Rev. Mol. Cell Biol. 2, 237–246 (2001)
Calvo, O. & Manley, J. L. Strange bedfellows: polyadenylation factors at the promoter. Genes Dev. 17, 1321–1327 (2003)
Vinciguerra, P. & Stutz, F. mRNA export: an assembly line from genes to nuclear pores. Curr. Opin. Cell Biol. 16, 285–292 (2004)
Shatkin, A. J. & Manley, J. L. The ends of the affair: capping and polyadenylation. Nature Struct. Biol. 7, 838–842 (2000)
Wickens, M. & Gonzalez, T. N. Knives, accomplices, and RNA. Science 306, 1299–1300 (2004)
Daiyasu, H., Osaka, K., Ishino, Y. & Toh, H. Expansion of the zinc metallo-hydrolase family of the β-lactamase fold. FEBS Lett. 503, 1–6 (2001)
Callebaut, I., Moshous, D., Mornon, J.-P. & de Villartay, J.-P. Metallo-β-lactamase fold within nucleic acids processing enzymes: the β-CASP family. Nucleic Acids Res. 30, 3592–3601 (2002)
Ryan, K., Calvo, O. & Manley, J. L. Evidence that polyadenylation factor CPSF-73 is the mRNA 3' processing endonuclease. RNA 10, 565–573 (2004)
Dominski, Z., Yang, X.-C. & Marzluff, W. F. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123, 37–48 (2005)
Kolev, N. G. & Steitz, J. A. Symplekin and multiple other polyadenylation factors participate in 3'-end maturation of histone mRNAs. Genes Dev. 19, 2583–2592 (2005)
Mandel, C. R., Gebauer, D., Zhang, H. & Tong, L. A serendipitous discovery that in situ proteolysis is required for the crystallization of yeast CPSF-100 (Ydh1p). Acta Crystallogr. F62, 1041–1045 (2006)
Ullah, J. H. et al. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7Å resolution. J. Mol. Biol. 284, 125–136 (1998)
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993)
de la Sierra-Gallay, I. L., Pellegrini, O. & Condon, C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature 433, 657–661 (2005)
Ishii, R. et al. Crystal structure of the tRNA 3' processing endoribonuclease tRNase Z from Thermotoga maritima.. J. Biol. Chem. 280, 14138–14144 (2005)
Walker, J. E., Saraste, M., Runswick, M. J. & Gay, N. J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 (1982)
Hirose, Y. & Manley, J. L. Creatine phosphate, not ATP, is required for 3' end cleavage of mammalian pre-mRNA in vitro.. J. Biol. Chem. 272, 29636–29642 (1997)
Garrity, J. D., Bennett, B. & Crowder, M. W. Direct evidence that the reaction intermediate of metallo-β-lactamase L1 is metal bound. Biochemistry 44, 1078–1087 (2005)
Baillat, D. et al. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123, 265–276 (2005)
Dominski, Z., Yang, X.-C., Purdy, M., Wagner, E. J. & Marzluff, W. F. A. CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol. 25, 1489–1500 (2005)
Moshous, D. et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105, 177–186 (2001)
Takagaki, Y., Ryner, L. C. & Manley, J. L. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell 52, 731–742 (1988)
Ma, Y., Pannicke, U., Schwarz, K. & Lieber, M. R. Hairpin opening and overhand processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108, 781–794 (2002)
Hendrickson, W. A. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science 254, 51–58 (1991)
Carson, M. Ribbon models of macromolecules. J. Mol. Graph. 5, 103–106 (1987)
Acknowledgements
We thank K. Ryan for discussions; B. Tweel for characterizing the fungus; R. Abramowitz, J. Schwanof and X. Yang for setting up the X4A beamline at the NSLS; and J. Khan and Y. Shen for help with data collection at the synchrotron. This research is supported in part by grants from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The atomic coordinates have been deposited at the Protein Data Bank (accession numbers 2I7T, 2I7V and 2I7X). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Results, Supplementary Methods, Supplementary Tables 1 and 1 and Supplementary Figures 1–9. (PDF 3416 kb)
Rights and permissions
About this article
Cite this article
Mandel, C., Kaneko, S., Zhang, H. et al. Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease. Nature 444, 953–956 (2006). https://doi.org/10.1038/nature05363
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05363
This article is cited by
-
The anticancer compound JTE-607 reveals hidden sequence specificity of the mRNA 3′ processing machinery
Nature Structural & Molecular Biology (2023)
-
Genomic regulation of transcription and RNA processing by the multitasking Integrator complex
Nature Reviews Molecular Cell Biology (2023)
-
Deep learning of human polyadenylation sites at nucleotide resolution reveals molecular determinants of site usage and relevance in disease
Nature Communications (2023)
-
1H, 15N and 13C resonance assignments of a minimal CPSF73-CPSF100 C-terminal heterodimer
Biomolecular NMR Assignments (2023)
-
Genome-wide screening for genetic variants in polyadenylation signal (PAS) sites in mouse selection lines for fatness and leanness
Mammalian Genome (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.