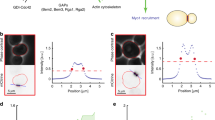
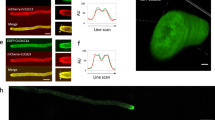
Abstract
Septins are polymerizing GTPases1 that function in cortical organization and cell division2,3,4. In Saccharomyces cerevisiae they localize at the isthmus between the mother and the daughter cells, where they undergo a transition from a non-dynamic hourglass-shaped assembly5 to two separate rings, at the onset of cytokinesis6,7. Septins form filaments as pure protein8 and in vivo9, but the filament organization within the hourglass and ring structures is controversial9,10. Here, we use polarized fluorescence microscopy11 of orientationally constrained green fluorescent protein to determine septin filament organization and dynamics in living yeast. We found that the hourglass is made of filaments aligned along the yeast bud neck. During the transition from hourglass to rings the filaments rotate through 90° in the membrane plane and become circumferential. These data resolve a long-standing controversy in the field and provide strong evidence that septins have a mechanical function in cell division.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Field, C. M. et al. A purified Drosophila septin complex forms filaments and exhibits GTPase activity. J. Cell Biol. 133, 605–616 (1996)
Kinoshita, M. et al. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 11, 1535–1547 (1997)
Neufeld, T. P. & Rubin, G. M. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell 77, 371–379 (1994)
Hartwell, L. H. Genetic control of the cell division cycle in yeast. IV. Genes controlling bud emergence and cytokinesis. Exp. Cell Res. 69, 265–276 (1971)
Dobbelaere, J. & Barral, Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science 305, 393–396 (2004)
Lippincott, J., Shannon, K. B., Shou, W., Deshaies, R. J. & Li, R. The Tem1 small GTPase controls actomyosin and septin dynamics during cytokinesis. J. Cell Sci. 114, 1379–1386 (2001)
Cid, V. J., Adamikova, L., Sanchez, M., Molina, M. & Nombela, C. Cell cycle control of septin ring dynamics in the budding yeast. Microbiology 147, 1437–1450 (2001)
Frazier, J. A. et al. Polymerization of purified yeast septins: evidence that organized filament arrays may not be required for septin function. J. Cell Biol. 143, 737–749 (1998)
Rodal, A. A., Kozubowski, L., Goode, B. L., Drubin, D. G. & Hartwig, J. H. Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell 16, 372–384 (2005)
Byers, B. & Goetsch, L. A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol. 69, 717–721 (1976)
Axelrod, D. Fluorescence polarization microscopy. Methods Cell Biol. 30, 333–352 (1989)
Sheff, M. A. & Thorn, K. S. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661–670 (2004)
Burghardt, T. P. Model-independent fluorescence polarization for measuring order in a biological assembly. Biopolymers 23, 2383–2406 (1984)
Dale, R. E. et al. Model-independent analysis of the orientation of fluorescent probes with restricted mobility in muscle fibers. Biophys. J. 76, 1606–1618 (1999)
Desper, C. R. & Kimura, I. Mathematics of the polarized-fluorescence experiment. J. Appl. Phys. 38, 4225–4233 (1967)
Inoue, S., Shimomura, O., Goda, M., Shribak, M. & Tran, P. T. Fluorescence polarization of green fluorescence protein. Proc. Natl Acad. Sci. USA 99, 4272–4277 (2002)
Axelrod, D. Carbocyanine dye orientation in red cell membrane studied by microscopic fluorescence polarization. Biophys. J. 26, 557–573 (1979)
Volkmer, A., Subramaniam, V., Birch, D. J. & Jovin, T. M. One- and two-photon excited fluorescence lifetimes and anisotropy decays of green fluorescent proteins. Biophys. J. 78, 1589–1598 (2000)
Corrie, J. E. et al. Dynamic measurement of myosin light-chain-domain tilt and twist in muscle contraction. Nature 400, 425–430 (1999)
Rocheleau, J. V., Edidin, M. & Piston, D. W. Intrasequence GFP in class I MHC molecules, a rigid probe for fluorescence anisotropy measurements of the membrane environment. Biophys. J. 84, 4078–4086 (2003)
Lupas, A., Van Dyke, M. & Stock, J. Predicting coiled coils from protein sequences. Science 252, 1162–1164 (1991)
Yang, F., Moss, L. G. & Phillips, G. N. Jr. The molecular structure of green fluorescent protein. Nature Biotechnol. 14, 1246–1251 (1996)
Ormo, M. et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273, 1392–1395 (1996)
Li, X. et al. Deletions of the Aequorea victoria green fluorescent protein define the minimal domain required for fluorescence. J. Biol. Chem. 272, 28545–28549 (1997)
Dobbelaere, J., Gentry, M. S., Hallberg, R. L. & Barral, Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev. Cell 4, 345–357 (2003)
Caviston, J. P., Longtine, M., Pringle, J. R. & Bi, E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14, 4051–4066 (2003)
Schmidt, M., Bowers, B., Varma, A., Roh, D. H. & Cabib, E. In budding yeast, contraction of the actomyosin ring and formation of the primary septum at cytokinesis depend on each other. J. Cell Sci. 115, 293–302 (2002)
Picart, C. & Discher, D. E. Actin protofilament orientation at the erythrocyte membrane. Biophys. J. 77, 865–878 (1999)
Tatchell, K. & Robinson, L. C. Use of green fluorescent protein in living yeast cells. Methods Enzymol. 351, 661–683 (2002)
Acknowledgements
We thank I. Vrabioiu and colleagues at INCDMF-CEFIN, Romania, for designing and fabricating the rotating stage used for our experiments, and M. Volles for comments. This work was supported by a National Institutes of Health grant to T.J.M.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Methods, Supplementary Discussion, Supplementary Notes, Supplementary Tables, Supplementary Figures and Supplementary Movie Legend. (DOC 1163 kb)
Supplementary Figure 1
Characterization of the Cdc12 strain. (JPG 24 kb)
Supplementary Figure 2
Hourglass area C quantitations. (JPG 17 kb)
Supplementary Figure 3
Cdc3 strain rearrangement time lapse. (JPG 22 kb)
Supplementary Movie
This movie indicates that the septin filaments change orientation during the hourglass to rings transition. (MOV 4949 kb)
Rights and permissions
About this article
Cite this article
Vrabioiu, A., Mitchison, T. Structural insights into yeast septin organization from polarized fluorescence microscopy. Nature 443, 466–469 (2006). https://doi.org/10.1038/nature05109
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05109
This article is cited by
-
Polarization modulation with optical lock-in detection reveals universal fluorescence anisotropy of subcellular structures in live cells
Light: Science & Applications (2022)
-
Membrane reshaping by micrometric curvature sensitive septin filaments
Nature Communications (2019)
-
Super-resolution imaging of fluorescent dipoles via polarized structured illumination microscopy
Nature Communications (2019)
-
The anillin-related Int1 protein and the Sep7 septin collaborate to maintain cellular ploidy in Candida albicans
Scientific Reports (2018)
-
Recruitment of the mitotic exit network to yeast centrosomes couples septin displacement to actomyosin constriction
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.