Abstract
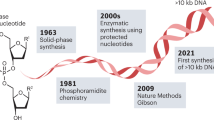
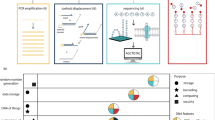
Testing the many hypotheses from genomics and systems biology experiments demands accurate and cost-effective gene and genome synthesis. Here we describe a microchip-based technology for multiplex gene synthesis. Pools of thousands of ‘construction’ oligonucleotides and tagged complementary ‘selection’ oligonucleotides are synthesized on photo-programmable microfluidic chips1, released, amplified and selected by hybridization to reduce synthesis errors ninefold. A one-step polymerase assembly multiplexing reaction assembles these into multiple genes. This technology enabled us to synthesize all 21 genes that encode the proteins of the Escherichia coli 30S ribosomal subunit, and to optimize their translation efficiency in vitro through alteration of codon bias. This is a significant step towards the synthesis of ribosomes in vitro and should have utility for synthetic biology in general.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Zhou, X. et al. Microfluidic PicoArray synthesis of oligodeoxynucleotides and simultaneous assembling of multiple DNA sequences. Nucleic Acids Res. 32, 5409–5417 (2004)
Fodor, S. P. et al. Light-directed, spatially addressable parallel chemical synthesis. Science 251, 767–773 (1991)
Gao, X. et al. Flexible DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res. 29, 4744–4750 (2001)
Cello, J., Paul, A. V. & Wimmer, E. Chemical synthesis of poliovirus cDNA: generation of infectious virus in the absence of natural template. Science 297, 1016–1018 (2002)
Smith, H. O., Hutchison, C. A. III, Pfannkoch, C. & Venter, J. C. Generating a synthetic genome by whole genome assembly: ΦX174 bacteriophage from synthetic oligonucleotides. Proc. Natl Acad. Sci. USA 100, 15440–15445 (2003)
Stemmer, W. P., Crameri, A., Ha, K. D., Brennan, T. M. & Heyneker, H. L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164, 49–53 (1995)
Eason, R. G. et al. Characterization of synthetic DNA bar codes in Saccharomyces cerevisiae gene-deletion strains. Proc. Natl Acad. Sci. USA 101, 11046–11051 (2004)
Forster, A. C. & Church, G. M. A synthetic biology project. Nature (submitted)
Culver, G. M. & Noller, H. F. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA 5, 832–843 (1999)
Iost, I., Guillerez, J. & Dreyfus, M. Bacteriophage T7 RNA polymerase travels far ahead of ribosomes in vivo. J. Bacteriol. 174, 619–622 (1992)
Iost, I. & Dreyfus, M. mRNAs can be stabilized by DEAD-box proteins. Nature 372, 193–196 (1994)
Carr, P. A., Park, J. S., Lee, Y.-J., Yu, T., Zhang, S. & Jacobson, J. M. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res. (in the press)
Smith, J. & Modrich, P. Removal of polymerase-produced mutant sequences from PCR products. Proc. Natl Acad. Sci. USA 94, 6847–6850 (1997)
Shendure, J., Mitra, R., Varma, C. & Church, G. M. Advanced sequencing technologies: methods and goals. Nature Rev. Genet. 5, 335–344 (2004)
Mullis, K. et al. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. LI, 263–273 (1986)
Dillon, P. J. & Rosen, C. A. A rapid method for the construction of synthetic genes using the polymerase chain reaction. BioTechniques 9, 298–300 (1990)
Liu, D., Park, S. H., Reif, J. H. & LaBean, T. H. DNA nanotubes self-assembled from triple-crossover tiles as templates for conductive nanowires. Proc. Natl Acad. Sci. USA 101, 717–722 (2004)
Jaffe, J. D., Berg, H. C. & Church, G. M. Proteogenomic mapping reveals genomic structure and novel proteins undetected by computational algorithms. Proteomics 4, 59–77 (2004)
Breslauer, K. J., Frank, R., Blöcker, H. & Marky, L. A. Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA 83, 3746–3750 (1986)
Richmond, K. E. et al. Amplification and assembly of chip-eluted DNA (AACED): a method for high-throughput gene synthesis. Nucleic Acids Res. 32, 5011–5018 (2004)
Espelund, M., Stacy, R. A. & Jakobsen, K. S. A simple method for generating single-stranded DNA probes labeled to high activities. Nucleic Acids Res. 18, 6157–6158 (1990)
Acknowledgements
DARPA BioComp and DOE GTL provided support for J.T., H.G. and G.C. We thank G. Culver, T. Wu, T. Forster, P. Carr, J. Jacobson and other members of the synthetic biology community for advice; N. Novikov for technical assistance, and E. Nuwaysir and T. Albert for help in designing the Nimblegen arrays. J.T. was supported by a LSRF fellowship. X.Z., E.G. and X.G. thank the NIH, DARPA and the R.A. Welch Foundation for grants.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
X.Z., E.G. and X.G. have potentially competing financial interests in Atactic Technologies. G.C. and J.T. have potentially competing financial interests in SynBio Corp.
Supplementary information
Supplementary Table 1 and 2
Table 1: Sequences of the construction oligonucleotides, selection oligonucleotides, oligonucleotide PCR-amplification primers, and gene-end primers; Table 2: Primers for adding His-tags, linkers to construct overlaps, 3 secondary assembly primers, and 1 final PCR assembly flanking primers. (DOC 136 kb)
Supplementary Methods
Description of methods for making the synthetic 14,593-nt operon of 21 codon-modified E.coli 30S ribosomal genes and lists of oligonucleotides used. (DOC 24 kb)
Rights and permissions
About this article
Cite this article
Tian, J., Gong, H., Sheng, N. et al. Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432, 1050–1054 (2004). https://doi.org/10.1038/nature03151
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03151
This article is cited by
-
Catalytic length-controlled oligomerization with synthetic programmable templates
Nature Synthesis (2023)
-
DNA synthesis technologies to close the gene writing gap
Nature Reviews Chemistry (2023)
-
Catalytic cavities control oligomer length
Nature Synthesis (2023)
-
Numerical Simulation of Mixing in Active Micromixers Using SPH
Transport in Porous Media (2023)
-
Purification of multiplex oligonucleotide libraries by synthesis and selection
Nature Biotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.