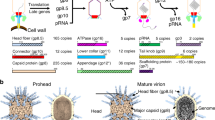
Abstract
The structure of the membrane-containing bacteriophage PRD1 has been determined by X-ray crystallography at about 4 Å resolution. Here we describe the structure and location of proteins P3, P16, P30 and P31. Different structural proteins seem to have specialist roles in controlling virus assembly. The linearly extended P30 appears to nucleate the formation of the icosahedral facets (composed of trimers of the major capsid protein, P3) and acts as a molecular tape-measure, defining the size of the virus and cementing the facets together. Pentamers of P31 form the vertex base, interlocking with subunits of P3 and interacting with the membrane protein P16. The architectural similarities with adenovirus and one of the largest known virus particles PBCV-1 support the notion that the mechanism of assembly of PRD1 is scaleable and applies across the major viral lineage formed by these viruses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bamford, J. K. et al. Diffraction quality crystals of PRD1, a 66-MDa dsDNA virus with an internal membrane. J. Struct. Biol. 139, 103–112 (2002)
Rydman, P. S. et al. Bacteriophage PRD1 contains a labile receptor-binding structure at each vertex. J. Mol. Biol. 291, 575–587 (1999)
Rydman, P. S., Bamford, J. K. & Bamford, D. H. A minor capsid protein P30 is essential for bacteriophage PRD1 capsid assembly. J. Mol. Biol. 313, 785–795 (2001)
Mindich, L., Bamford, D., McGraw, T. & Mackenzie, G. Assembly of bacteriophage PRD1: particle formation with wild-type and mutant viruses. J. Virol. 44, 1021–1030 (1982)
Stromsten, N. J., Bamford, D. H. & Bamford, J. K. The unique vertex of bacterial virus PRD1 is connected to the viral internal membrane. J. Virol. 77, 6314–6321 (2003)
Gowen, B., Bamford, J. K., Bamford, D. H. & Fuller, S. D. The tailless icosahedral membrane virus PRD1 localizes the proteins involved in genome packaging and injection at a unique vertex. J. Virol. 77, 7863–7871 (2003)
Bamford, D. H., Caldentey, J. & Bamford, J. K. Bacteriophage PRD1: a broad host range dsDNA tectivirus with an internal membrane. Adv. Virus Res. 45, 281–319 (1995)
Grahn, A. M., Daugelavicius, R. & Bamford, D. H. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol. Microbiol. 46, 1199–1209 (2002)
Grahn, A. M., Daugelavicius, R. & Bamford, D. H. The small viral membrane-associated protein P32 is involved in bacteriophage PRD1 DNA entry. J. Virol. 76, 4866–4872 (2002)
Butcher, S. J., Bamford, D. H. & Fuller, S. D. DNA packaging orders the membrane of bacteriophage PRD1. EMBO J. 14, 6078–6086 (1995)
Benson, S. D., Bamford, J. K., Bamford, D. H. & Burnett, R. M. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell 98, 825–833 (1999)
Belnap, D. M. & Steven, A. C. ‘Deja vu all over again’: the similar structures of bacteriophage PRD1 and adenovirus. Trends Microbiol. 8, 91–93 (2000)
Bamford, D. H., Burnett, R. M. & Stuart, D. I. Evolution of viral structure. Theor. Popul. Biol. 61, 461–470 (2002)
Yan, X. et al. Structure and assembly of large lipid-containing dsDNA viruses. Nature Struct. Biol. 7, 101–103 (2000)
Nandhagopal, N. et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl Acad. Sci. USA 99, 14758–14763 (2002)
San Martín, C. et al. Combined EM/X-ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1 and shows key capsid and membrane interactions. Structure 9, 917–930 (2001)
San Martín, C. et al. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nature Struct. Biol. 9, 756–763 (2002)
Cockburn, J. J., Bamford, J. K., Grimes, J. M., Bamford, D. H. & Stuart, D. I. Crystallization of the membrane-containing bacteriophage PRD1 in quartz capillaries by vapour diffusion. Acta Crystallogr. D 59, 538–540 (2003)
Cockburn, J. J. et al. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature doi:10.1038/nature03053 (this issue)
Benson, S. D., Bamford, J. K., Bamford, D. H. & Burnett, R. M. The X-ray crystal structure of P3, the major coat protein of the lipid-containing bacteriophage PRD1, at 1.65 Å resolution. Acta Crystallogr. D 58, 39–59 (2002)
Li, X., Romero, P., Rani, M., Dunker, A. K. & Obradovic, Z. Predicting protein disorder for N-, C-, and internal regions. Genome Informatics 10, 30–40 (1999)
Stuart, D. I., Levine, M., Muirhead, H. & Stammers, D. K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol. 134, 109–142 (1979)
Liddington, R. C. et al. Structure of simian virus 40 at 3.8 Å resolution. Nature 354, 278–284 (1991)
Grimes, J. M. et al. The atomic structure of the bluetongue virus core. Nature 395, 470–478 (1998)
Chiu, W., Burnett, R. M. & Garcea, R. L. in Structural Biology of Viruses (eds Chiu, W., Burnett, R. M. & Garcea, R. L.) 209–238 (Oxford Univ. Press, New York, 1997)
Caldentey, J., Tuma, R. & Bamford, D. H. Assembly of bacteriophage PRD1 spike complex: role of the multidomain protein P5. Biochemistry 39, 10566–10573 (2000)
Sokolova, A. et al. Solution structure of bacteriophage PRD1 vertex complex. J. Biol. Chem. 276, 46187–46195 (2001)
Xu, L., Benson, S. D., Butcher, S. J., Bamford, D. H. & Burnett, R. M. The receptor binding protein P2 of PRD1, a virus targeting antibiotic-resistant bacteria, has a novel fold suggesting multiple functions. Structure 11, 309–322 (2003)
Jaatinen, S., Viitanen, S., Bamford, D. H. & Bamford, J. K. The integral membrane protein P16 of bacteriophage PRD1 stabilizes the adsorption vertex structure. J. Virol. 78, 9790–9797 (2004)
Luo, C., Butcher, S. & Bamford, D. H. Isolation of a phospholipid-free protein shell of bacteriophage PRD1, an Escherichia coli virus with an internal membrane. Virology 194, 564–569 (1993)
Stewart, P. L., Fuller, S. D. & Burnett, R. M. Difference imaging of adenovirus: bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 12, 2589–2599 (1993)
Bamford, D. & Mindich, L. Structure of the lipid-containing bacteriophage PRD1: disruption of wild-type and nonsense mutant phage particles with guanidine hydrochloride. J. Virol. 44, 1031–1038 (1982)
Bamford, J. K. et al. Genome organization of membrane-containing bacteriophage PRD1. Virology 183, 658–676 (1991)
Rydman, P. S. & Bamford, D. H. Bacteriophage PRD1 DNA entry uses a viral membrane-associated transglycosylase activity. Mol. Microbiol. 37, 356–363 (2000)
Rydman, P. S. & Bamford, D. H. The lytic enzyme of bacteriophage PRD1 is associated with the viral membrane. J. Bacteriol. 184, 104–110 (2002)
Thuman-Commike, P. A. et al. Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids. Biophys. J. 76, 3267–3277 (1999)
Morais, M. C. et al. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nature Struct. Biol. 10, 572–576 (2003)
Journet, L., Agrain, C., Broz, P. & Cornelis, G. R. The needle length of bacterial injectisomes is determined by a molecular ruler. Science 302, 1757–1760 (2003)
Burnett, R. M. The structure of the adenovirus capsid. II. The packing symmetry of hexon and its implications for viral architecture. J. Mol. Biol. 185, 125–143 (1985)
Sambrook, J. & Russell, D. W. Molecular Cloning: a Laboratory Manual 3rd edn (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, 2001)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, Number 4. The CCP 4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Brunger, A. T. X-PLOR, Version 3.1: A system for X-ray crystallography and NMR (Yale Univ. Press, New Haven, Connecticut, 1992)
DeLano, W. L. & Brunger, A. T. The direct rotation function – rotational Patterson correlation search applied to molecular replacement. Acta Crystallogr. D 51, 740–748 (1995)
Diprose, J. M. et al. Translocation portals for the substrates and products of a viral transcription complex: the bluetongue virus core. EMBO J. 20, 7229–7239 (2001)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Sonnhammer, E. L. L., von Heijne, G. & Krogh, A. in Proc. Sixth International Conference on Intelligent Systems for Molecular Biology (ed. Glasgow, J.) 175–182 (AAAI Press, Menlo Park, California, 1998)
Esnouf, R. M. Polyalanine reconstruction from Cα positions using the program CALPHA can aid initial phasing of data by molecular replacement procedures. Acta Crystallogr. D 53, 665–672 (1997)
Frishman, D. & Argos, P. Seventy-five percent accuracy in protein secondary structure prediction. Proteins 27, 329–335 (1997)
Acknowledgements
We thank M.-L. Perälä and S. Ollila for technical assistance, the staff at the European Synchrotron Radiation Facility for beamline support, and E. Mancini for assistance with data collection. This investigation was supported by research grants from the Academy of Finland to J.K.H.B. and S.J.B., research grants and the Finnish Centre of Excellence Program (2000–2005) from the Academy of Finland to D.H.B., a grant from the National Science Foundation to R.M.B., the Biotechnology and Biological Sciences Research Council and the Medical Research Council, UK and a grant from the Human Frontiers Science Program to D.I.S., R.M.B. and D.H.B. S.D.F. is supported by the Wellcome Trust, J.M.G. by the Royal Society and D.I.S. by the UK Medical Research Council.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure
Curves of data completeness and correlation coefficient versus resolution for cell1 and cell2 data. (DOC 416 kb)
Supplementary Methods
Describes the calculation of packing density in the cavity under the vertex. (DOC 24 kb)
Rights and permissions
About this article
Cite this article
Abrescia, N., Cockburn, J., Grimes, J. et al. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432, 68–74 (2004). https://doi.org/10.1038/nature03056
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03056
This article is cited by
-
Rapid, sensitive, and low-cost detection of Escherichia coli bacteria in contaminated water samples using a phage-based assay
Scientific Reports (2022)
-
Cryo-EM structure of ssDNA bacteriophage ΦCjT23 provides insight into early virus evolution
Nature Communications (2022)
-
Phage diversity, genomics and phylogeny
Nature Reviews Microbiology (2020)
-
Structural basis for assembly of vertical single β-barrel viruses
Nature Communications (2019)
-
Near-atomic structure of a giant virus
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.