Abstract
Cryptosporidium species cause acute gastroenteritis and diarrhoea worldwide. They are members of the Apicomplexa—protozoan pathogens that invade host cells by using a specialized apical complex and are usually transmitted by an invertebrate vector or intermediate host. In contrast to other Apicomplexans, Cryptosporidium is transmitted by ingestion of oocysts and completes its life cycle in a single host. No therapy is available, and control focuses on eliminating oocysts in water supplies1. Two species, C. hominis and C. parvum, which differ in host range, genotype and pathogenicity, are most relevant to humans1,2,3. C. hominis is restricted to humans, whereas C. parvum also infects other mammals2. Here we describe the eight-chromosome ∼9.2-million-base genome of C. hominis2. The complement of C. hominis protein-coding genes shows a striking concordance with the requirements imposed by the environmental niches the parasite inhabits. Energy metabolism is largely from glycolysis. Both aerobic and anaerobic metabolisms are available, the former requiring an alternative electron transport system in a simplified mitochondrion. Biosynthesis capabilities are limited, explaining an extensive array of transporters. Evidence of an apicoplast is absent, but genes associated with apical complex organelles are present. C. hominis and C. parvum exhibit very similar gene complements, and phenotypic differences between these parasites must be due to subtle sequence divergence.
Similar content being viewed by others
Main
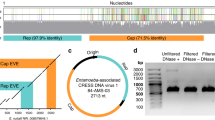
We generated a ∼12-fold sequence and ∼8-fold bacterial artificial chromosome (BAC) clone coverage of the genome of C. hominis isolate TU502 (ref. 3, Fig. 1, Supplementary Figs 1–8, Supplementary Tables 1 and 2). Alignment of the ∼9.2-million-base (Mb) final sequence with the HAPPY map4 and chromosomes of the C. parvum genome5 covered ∼9.1 Mb. The eight chromosomes range from ∼0.9 to ∼1.4 Mb and exhibit 31.7% GC content (compare with 30.3% and 19.4% for C. parvum and P. falciparum6, respectively). The density of 2–50-base-pair (bp) repeats was about 1 per 2,800 bp. The distribution of repeats is biased towards chromosome ends because over 85% are in the telomere-proximal thirds of five of the chromosomes (Supplementary Fig. 9). Two octamers, TGGCGCCA and TGCATGCA, over-represented in other apicomplexans4, are ∼40-fold and 15-fold over-represented in C. hominis (Supplementary Table 3). More than 80% of these are in non-coding sequences, indicating possible regulatory or other conserved function. Forty-five tRNAs, four or five rRNA operons—at least one of each of the two known types (Supplementary Table 4)—and two clusters of three tandem 5S rRNA genes are present. As in P. falciparum6, two tRNAMet genes are present, suggesting discrete roles in initiation and extension. We estimate that there are ∼3,994 genes in C. hominis, in comparison with 3,952 genes in C. parvum and 5,268 in P. falciparum6 (Table 1). About 60% exhibit similarity to known genes. The distribution of GO annotations for Cryptosporidium, Plasmodium and Saccharomyces, is remarkably similar (Supplementary Fig. 10), indicating that their phenotypic differences are a reflection of non-conserved or previously unreported gene families of unknown function rather than to the functional specialization of conserved gene families. We estimate that 5–20% of C. hominis genes have introns.
Tracks indicate C. hominis contigs (blue), sequence gaps (white) and physical gaps (red) (a); HAPPY4 markers (b); positions of the octamers TGGCGCCA (c) and TGCATGCA (d); Gene Ontology (GO) of molecular function for the predicted genes shown by strand (see key, left (e) and right (f)); tRNAs (blue) and rRNAs (magenta) (g); percentage identity to C. parvum in 5-kb windows (see key; average identities are shown at the foot of each chromosome) (h); BAC clone coverage (overlapping clones collapsed to a single line) (i). The scale to the left of each chromosome represents C. parvum sequences (red triangles show sequence gaps), with the first base at the top.
Analysis of the C. hominis genome shows that the parasite possesses a highly tailored glycolysis-based metabolism, is dependent on the host for nutrients, and is exquisitely adapted for its life cycle (Fig. 2, Supplementary Tables 5 and 6). Glycolysis seems functional, unlike the tricarboxylic acid (TCA) cycle and oxidative phosphorylation. Both an anaerobic pathway using pyruvate:NADP+ oxidoreductase (PNO) and an aerobic pathway using an alternative oxidase (AOX) are available for recycling NAD+ to NADH. In the former, pyruvate is fermented to acetyl coenzyme A (acetyl-CoA) producing NADPH, which is then reduced to NADP+, releasing hydrogen, by a Narf-like [Fe]-hydrogenase, as in Trichomonas7. Acetyl-CoA is processed by acetate CoA synthase to produce acetate and ATP, as in Giardia8, yielding four ATP per glucose. Acetyl-CoA can also be processed to ethanol yielding no additional ATP. Under glucose-limited conditions, conversion of acetyl-CoA to acetate, generating two extra ATP per glucose, might be favoured. When glucose is in excess, pyruvate can be converted to lactate or ethanol to regenerate NAD+ but no additional ATP. C. hominis can also generate ATP by metabolism of glycerol using glycerol-3-phosphate dehydrogenase and triose phosphate isomerase.
The green strip represents the cellular membrane with putative transporters; numbers indicate the number of genes for a given class of transporter. Solid arrows indicate pathways that are present; multistep pathways are indicated with dashed arrows. Components or pathways that are absent are crossed out. Steps or components whose exact nature is questionable are shown with question marks. Blue arrows and names indicate proposed aerobic parts of the metabolism. Abbreviations: ABC, ATP-binding cassette; AC, adenylyl cyclase; Ado, adenosine; AOX, alternative oxidase; Cpn60, chaperone 60; Cyd, cytidine; DHF, dihydrofolate; dThd, deoxythymidine; GPI, glycosylphosphatidylinositol; Hsp70, heat-shock protein 70; InsP3, inositol phosphate; MRP, multiple-drug-resistance protein; NADH DH, NADH dehydrogenase; Narf-like, nuclear prelamin A recognition factor-like protein; PEP, phosphoenolpyruvate; PI(3)K, phosphatidylinositol 3-kinase; PKA, protein kinase A; PLC, phospholipase C; PKC, protein kinase C; PNO–CPR, pyruvate:NADP+ oxidoreductase fused to cytochrome P450 reductase domain; THF, tetrahydrofolate; TIM17, translocase of the inner mitochondrial membrane 17; TOM40, translocase of the outer mitochondrial membrane 40; UQ, ubiquinone; Urd, uridine.
C. hominis can convert pyruvate to malate and subsequently to oxaloacetate (OAA), regenerating NAD+. However, malate shuttle enzymes—for example, aspartate amino transferase—which process OAA to aspartic acid for export from the mitochondrion, are absent. Cytoplasmic malate could be converted to OAA by a mitochondrial membrane-bound malate dehydrogenase, like the lactate shuttle of Euglena gracilis9, passing electrons from malate to an electron transport system composed of elements of Complexes I and III and an alternative oxidase system with O2 as electron acceptor and producing no additional ATP.
Enzymes for metabolism of glycogen, starch and amylopectin are present, which is consistent with suggestions that amylopectin represents an energy reserve for sporozoites10. Lack of glucose-6-phosphate 1-dehydrogenase and other enzymes of the pentose phosphate pathway suggests that, unlike P. falciparum and other apicomplexans6, C. hominis cannot metabolize five-carbon sugars or nucleotides. Components of β-oxidation, for example enoyl-CoA hydratase and acetyl-CoA C-acyltransferase, are also absent, precluding ATP generation from fatty acids. Enzymes for the catabolism of proteins are also absent.
Major TCA-cycle enzymes—isocitrate dehydrogenase, succinyl-CoA synthase and succinate dehydrogenase—are absent in C. hominis. Despite the presence of ubiquinol-cytochrome c reductase, NADH dehydrogenase (ubiquinone), H+-transporting ATPase and iron–sulphur cluster-like proteins, among others, key components of Complexes II and IV are absent, precluding ATP generation by oxidative phosphorylation. Components of oxidative phosphorylation that are present (parts of Complexes I and III) probably reoxidize NADH in a simplified electron-transport chain, as in some plants and protozoa.
Consistent with previous suggestions11 is the observation that Cryptosporidium lacks enzymes for the synthesis of key biochemical building blocks—simple sugars, amino acids and nucleotides. However, starch, amylopectin and fatty acids can be generated from precursors. Interestingly, these C. hominis enzymes have minimal similarity to the known biosynthetic enzymes and are potential therapeutic targets.
Enzymes of the TCA, urea and nitrogen cycles and of the shikimate pathway are absent, indicating that Cryptosporidium is an amino-acid auxotroph. The shikimate pathway has been proposed as a potential target for glyphosate-based chemotherapy in other parasites including Cryptosporidium. We found no evidence to support this hypothesis. Enzymes that interconvert amino acids are encoded in C. hominis, and, unlike P. falciparum6, C. hominis has a large complement of amino acid transporters.
C. hominis lacks enzymes to synthesize bases or nucleosides, but encodes enzymes that convert nucleosides into nucleotides and interconvert nucleotides. As in other parasites, thymidylate synthase and dihydrofolate reductase of C. hominis are encoded as a bifunctional polypeptide, and novel polymorphisms at crucial sites have been proposed to explain Cryptosporidium's resistance to antifolates12. As previously suggested11, several nucleotide conversion enzymes seem to have a prokaryotic origin.
Fatty-acid biosynthesis in apicomplexans occurs in the apicoplast by means of a type II system including fatty-acid synthase (FAS). However, consistent with the absence of an apicoplast in Cryptosporidium13 is the observation that C. hominis encodes large FAS and polyketide synthase (PKS) enzymes, indicating a type I mechanism. The type I FAS and PKS enzymes of C. hominis also have prokaryotic characteristics14,15.
Glycerolipid and phospholipid metabolic pathways for phosphatidylinositol biosynthesis are available in C. hominis. 1,2-Diacylglycerol is a precursor for glycosylphosphatidylinositol anchor synthesis. All enzymes required for synthesis of these anchors are apparently present16.
Polyamines like putrescine, spermine and spermidine are critical for cellular viability, and enzymes required for their synthesis are attractive therapeutic targets. Cryptosporidium can synthesize polyamines using arginine decarboxylase rather than ornithine decarboxylase17. The putative arginine decarboxylase, spermidine synthase and other relevant enzymes encoded by C. hominis have diverged significantly from their homologues and are potential therapeutic targets.
C. hominis encodes adenylate cyclase, cyclic-AMP phosphodiesterase and protein kinase A, indicating the presence of the cAMP-mediated signalling pathway (Supplementary Table 7). Trimeric G protein, often involved in the activation of cAMP-mediated signalling, was not found in C. hominis, indicating that, as in Kinetoplastida18 and reminiscent of plants, this pathway is independent of this complex in C. hominis. The presence of phosphatidylinositol 3-kinase and phospholipase C indicates that C. hominis utilizes phosphatidylinositol phosphate and Ca2+-mediated regulatory mechanisms. The presence of putative Ca2+ transporters, enzymes associated with acidocalcisomes, and calmodulin imply that Ca2+ transport and sequestering are functional. Protein kinase C receptors indicate that C. hominis has the ability to signal by activation of soluble cytoplasmic receptor-associated kinases.
No mitochondrial DNA sequences were found in C. hominis, and the TCA cycle and oxidative phosphorylation are absent (Supplementary Tables 5, 6 and 8). However, a double-membrane-bound organelle generates a proton gradient using cardiolipin and performs some related mitochondrial functions, and mitochondrial marker chaperonin 60 was localized to this structure19. Core enzymes of [Fe–S] cluster biosynthesis, namely CpFd1, IscU, IscS, mt-HSP70, mtFNR and frataxin, have been reported in Cryptosporidium20, and we were not surprised to observe proteins involved in electron transport. We used CDART21 to identify [Fe–S] domains in HscB (JAC) and ATM1, which are possibly involved in chaperonin activity of Hsp40/DnaJ type and ABC transport. Thus, C. hominis, like the microsporidian Encephalitozoon cuniculi22, another obligate intracellular parasite, contains a minimal set of these proteins. These results imply significant mitochondrial function in C. hominis and indicate that the previously reported organelle19 is an atypical mitochondrion.
Cryptosporidium apparently lacks an apicoplast13,23, and searches of the C. hominis genome identified no apicoplast-encoded genes (Supplementary Table 9). Some putative nuclear-encoded apicoplast genes, for example acetyl-CoA carboxylase 1 precursor24 and adenylyl cyclase25, are present. Others, such as the apicoplast 50S ribosomal protein L33 and the ribosomal L28 and S9 precursor proteins, were not found. The data indicate that Cryptosporidium lost an ancestral apicoplast. The presence of d-glucose-6-phosphate ketol-isomerase and 2-phospho-d-glycerate hydrolase, which are similar to plant genes and may be derived from ancient algal endosymbionts, is also indicative that engulfment of the alga that gave rise to the apicoplast preceded the divergence of Cryptosporidium from other apicomplexans. One hypothesis is that the acquisition of the type 1 FAS by a progenitor organism obviated the fatty-acid synthesis capabilities of the apicoplast14,15.
The C. hominis genome encodes multiple proteins specific for components of the apical complex including micronemes and rhoptries (Supplementary Table 9). No specific dense granule-associated proteins were observed, probably because these proteins diverge rapidly26. However, proteins implicated in the regulation of transport and enhancement of the release of dense granule proteins27 are present. As for Plasmodium, a typical Golgi structure is not apparent in C. hominis23. However, the presence of secretory organelles implies the existence of a functional endoplasmic reticulum and Golgi, and C. hominis encodes proteins similar to many related components, including the NSF/SNAP/SNARE/Rab machinery, which participates in dense granule release28, and the rhoptry biogenesis mediator activator protein 1, involved in endoplasmic-reticulum–Golgi-organelle protein traffic29. The endoplasmic-reticulum–Golgi-organelle machinery of C. hominis therefore seems similar to that of other apicomplexans.
As described above, C. hominis exhibits limited biosynthetic capabilities and is apparently dependent on its ability to import essential nutrients such as amino acids, nucleotides and simple sugars. The genome encodes more than 80 genes with strong similarity to known transporters and several hundred genes with transporter-like properties. At least 12 sugar or nucleotide-sugar transporters, five putative amino-acid transporters, three fatty-acid transporters, 23 ABC family transporters including possible multiple-drug-resistance proteins, and several putative mitochondrial transporters are present. Other putative transporters for choline uptake, aminophospholipid transport, ATP/ADP, and others with unclear function, were also identified. These transporters are ideal therapeutic targets (Supplementary Table 10).
Comparison of the genomes of C. hominis and C. parvum (Fig. 1, Supplementary Table 11) showed that the two genomes are very similar, exhibiting only 3–5% sequence divergence with no large insertions, deletions (Supplementary Fig. 11) or rearrangements evident. In fact, the gene complements of the two species are essentially identical because the few C. parvum genes not found in C. hominis are proximal to known sequence gaps (Supplementary Table 1). We therefore conclude that the significant phenotypic differences between these parasites are due to functionally significant polymorphisms in relevant protein-coding genes and to subtle gene regulatory differences.
A striking feature of the C. hominis genome is the concordance between its gene complement and the metabolic requirements in the environmental niches of its two primary life-cycle stages—the quiescent oocyst in the nutrient-poor aerobic environment of contaminated water, and the vegetative parasites in the nutrient-rich anaerobic or microaerophilic environment of the host. Oocysts probably persist by aerobically metabolizing stores of complex carbohydrates by means of glycolysis and the alternative electron transport system in the unconventional mitochondrion. Consistent with the lack of the energy-generating TCA cycle, oxidative phosphorylation, β-oxidation and the pentose phosphate pathways is the observation that oocysts are relatively inactive, and the two ATP per glucose from glycolysis can provide sufficient energy. In the host, the parasite can import sugars to fuel glycolysis directly, netting two ATP per hexose. In limiting glucose, an additional two ATP per hexose can be generated either by converting acetyl-CoA to acetate or by means of glycerol metabolism. The residual mitochondrion lacks the TCA cycle and oxidative phosphorylation as expected in an organism that replicates in anaerobic or microaerophilic environments, and a simplified electron transport system for regenerating reducing power is available. Thus, a glycolysis-based metabolism is sufficient to support Cryptosporidium in all life-cycle stages.
As previously noted, our analysis shows that Cryptosporidium is a mosaic of sequences from diverse progenitors, including the hypothetical endosymbiont alga that formed the apicoplast, the mitochondrion and numerous genes acquired from prokaryotes by lateral transfer. Cryptosporidium also exhibits modular gene loss. We assume, on the basis of inference from other apicomplexans and earlier diverging groups such as the Euglenozoa, the Heterolobosea and the jakobids30, that Cryptosporidium progenitors exhibited the TCA cycle, β-oxidation, oxidative phosphorylation, amino acid, nucleotide and sugar biosynthesis, fully competent mitochondria, and a functional apicoplast. Genes associated with these functions are dispersed throughout the genome in Plasmodium and, we assume, in the progenitor. However, these systems seem to have been deleted cleanly in Cryptosporidium, leaving few identifiable residual genes or pseudogenes. Thus, the Cryptosporidium genome is a mosaic resulting from multiple lateral gene transfers and selective gene deletion.
The tailored physiology of C. hominis indicates attractive therapeutic targets (Supplementary Table 10), for example: essential transport systems; components of glycolysis; the unique prokaryotic FAS1 and PKS1; starch and amylopectin metabolism; nucleic-acid or amino-acid metabolism; the AOX electron transport system; the bifunctional thymidylate synthase–dihydrofolate reductase; and the diverged polyamine synthesis enzymes. Finally, many potential vaccine targets were identified in the C. hominis genome (not shown), and, in contrast with other protozoan parasites, no extensive arrays of potentially variant surface proteins were observed, indicating a possible role for immunoprophylaxis for cryptosporidiosis.
The availability of the genome sequence of the human pathogen C. hominis is a crucial step forward in our understanding of the biology of this parasite. The gene complement provides very significant insight into its physiology and metabolism, validating previous hypotheses and indicating the possibility of others. New obvious targets for chemotherapy and immunotherapy are already apparent. In short, we expect that the availability of the sequence of C. hominis will stimulate progress in research on this organism and its pathogenicity, and strategies for intervention in the diseases it causes.
Methods
A modified whole-genome shotgun strategy was used to sequence the ∼9.2-Mb genome of C. hominis isolate TU502, which was derived from an infected child from Uganda. DNA was purified from surface-sterilized oocysts, shotgun and BAC clones were constructed, and end sequences were generated. About 220,000 sequence reads from small insert clones, and end sequences from ∼2,000 BAC clones averaging ∼35 kbp in size, were generated. The data represents a ∼12-fold shotgun clone coverage of the genome with a quality score of Phred 20, and a 7–8-fold coverage with BAC clones. The sequences were assembled with Phrap, yielding a ∼9.16-Mb assembly, which was structurally and functionally analysed with a variety of available software programs and in-house scripts (see Supplementary Text 1 and 2 for further details and references).
References
Tzipori, S. & Ward, H. Cryptosporidiosis: biology, pathogenesis and disease. Microbes Infect. 4, 1047–1058 (2002)
Morgan-Ryan, U. M. et al. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49, 433–440 (2002)
Akiyoshi, D. E., Feng, X., Buckholt, M. A., Widmer, D. & Tzipori, S. Genetic analysis of Cryptosporidium parvum: human genotype 1 isolate passaged through different host species. Infect. Immun. 70, 5670–5675 (2002)
Bankier, A. T. et al. Integrated mapping, chromosomal sequencing and sequence analysis of Cryptosporidium parvum. Genome Res. 13, 1787–1799 (2003)
Abrahamsen, M. S. et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304, 441–445 (2004)
Gardner, M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002)
Muller, M. in Evolutionary Relationships among Protozoa (eds Coombs, G. H., Vickerman, K., Sleigh, M. A. & Warren, A.) 109–131 (Kluwer Academic, Dordrecht, 1998)
Sanchez, L. B., Galperin, M. Y. & Muller, M. Acetyl-CoA synthetase from the amitochondriate eukaryote Giardia lamblia belongs to the newly recognized superfamily of acyl-CoA synthetases (Nucleoside diphosphate-forming). J. Biol. Chem. 275, 5794–5803 (2000)
Jasso-Chavez, R. & Moreno-Sanchez, R. Cytosol-mitochondria transfer of reducing equivalents by a lactate shuttle in heterotrophic Euglena. Eur. J. Biochem. 270, 4942–4951 (2003)
Petry, F. & Harris, J. R. Ultrastructure, fractionation and biochemical analysis of Cryptosporidium parvum sporozoites. Int. J. Parasitol. 29, 1249–1260 (1999)
Striepen, B. et al. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc. Natl Acad. Sci. USA 101, 3154–3159 (2004)
Atreya, C. E. & Anderson, K. S. Kinetic characterization of bifunctional thymidylate synthase–dihydrofolate reductase (TS–DHFR) from Cryptosporidium hominis: A paradigm shift for TS activity and channeling behavior. J. Biol. Chem. 279, 18314–18322 (2004)
Zhu, G., Marchewka, M. J. & Keithly, J. S. Cryptosporidium parvum appears to lack a plastid genome. Microbiol. 146, 315–321 (2000)
Zhu, G. et al. Expression and functional characterization of a giant Type I fatty acid synthase (CpFAS1) gene from Cryptosporidium parvum. Mol. Biochem. Parasitol. 134, 127–135 (2004)
Zhu, G. et al. Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase. Gene 298, 79–89 (2002)
Priest, J. W., Xie, L. T., Arrowood, M. J. & Lammie, P. J. The immunodominant 17-kDa antigen from Cryptosporidium parvum is glycosylphosphatidylinositol-anchored. Mol. Biochem. Parasitol. 113, 117–126 (2001)
Keithly, J. S. et al. Polyamine biosynthesis in Cryptosporidium parvum and its implications for chemotherapy. Mol. Biochem. Parasitol. 88, 35–42 (1997)
Parsons, M. & Ruben, L. Pathways involved in environmental sensing in trypanosomatids. Parasitol. Today 16, 56–62 (2000)
Riordan, C. E., Ault, J. G., Langreth, S. G. & Keithly, J. S. Cryptosporidium parvum Cpn60 targets a relict organelle. Curr. Genet. 44, 138–147 (2003)
LaGier, M. J., Tachezy, J., Stejskal, F., Kutisova, K. & Keithly, J. S. Mitochondrial-type iron-sulfur cluster biosynthesis genes (IscS and IscU) in the apicomplexan Cryptosporidium parvum. Microbiol. 149, 3519–3530 (2003)
Geer, L. Y., Domrachev, M., Lipman, D. J. & Bryant, S. H. CDART: protein homology by domain architecture. Genome Res. 12, 1619–1623 (2002)
Katinka, M. D. et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414, 450–453 (2001)
Tetley, L., Brown, S. M., McDonald, V. & Coombs, G. H. Ultrastructural analysis of the sporozoite of Cryptosporidium parvum. Microbiol. 144, 3249–3255 (1998)
Zuther, E., Johnson, J. J., Haselkorn, R., McLeod, R. & Gornicki, P. Growth of Toxoplasma gondii is inhibited by aryloxyphenoxypropionate herbicides targeting acetyl-CoA carboxylase. Proc. Natl Acad. Sci. USA 96, 13387–13392 (1999)
Muhia, D. K. et al. Multiple splice variants encode a novel adenylyl cyclase of possible plastid origin expressed in the sexual stage of the malaria parasite Plasmodium falciparum. J. Biol. Chem. 278, 22014–22022 (2003)
Beyer, T. V., Svezhova, N. V., Radchenko, A. I. & Sidorenko, N. V. Parasitophorous vacuole: morphofunctional diversity in different coccidian genera (a short insight into the problem). Cell Biol. Int. 26, 861–871 (2002)
Stedman, T. T., Sussmann, A. R. & Joiner, K. A. Toxoplasma gondii Rab6 mediates a retrograde pathway for sorting of constitutively secreted proteins to the Golgi complex. J. Biol. Chem. 278, 5433–5443 (2003)
Chaturvedi, S. et al. Constitutive calcium-independent release of Toxoplasma gondii dense granules occurs through the NSF/SNAP/SNARE/Rab machinery. J. Biol. Chem. 274, 2424–2431 (1999)
Ngo, H. M. et al. AP-1 in Toxoplasma gondii mediates biogenesis of the rhoptry secretory organelle from a post-Golgi compartment. J. Biol. Chem. 278, 5343–5352 (2003)
Simpson, A. G. et al. Evolutionary history of ‘early-diverging’ eukaryotes: the excavate taxon Carpediemonas is a close relative of Giardia. Mol. Biol. Evol. 19, 1782–1791 (2002)
Acknowledgements
We thank S. Hendricks, M. R. C. Carvalho, S. Tula, G. Kazanina, J. Power, A. Holzgrefe, N. Ebashi, E. Butt, B. Sutton, S. Millett, W. Vogel and B. Constance for their technical contributions to this project and J. Elhai, D. Mallonee, L. M. Wen, T. Zwierzynski and Z. Chen for their contributions to the planning and performance of this project. This research was supported by grants from the National Institutes of Health.Author contributions S.T. and G.A.B. contributed equally.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Text 1
Methods and associated references. (DOC 40 kb)
Supplementary Text 2
Additional References. Important references not directly cited in the text of the manuscript. (DOC 55 kb)
Supplementary Figure 1
Schematic of C. hominis chromosome I. (PDF 186 kb)
Supplementary Figure 2
Schematic of C. hominis chromosome II. (PDF 194 kb)
Supplementary Figure 3
Schematic of C. hominis chromosome III. (PDF 195 kb)
Supplementary Figure 4
Schematic of C. hominis chromosome IV. (PDF 200 kb)
Supplementary Figure 5
Schematic of C. hominis chromosome V. (PDF 198 kb)
Supplementary Figure 6
Schematic of C. hominis chromosome VI. (PDF 218 kb)
Supplementary Figure 7
Schematic of C. hominis chromosome VII. (PDF 210 kb)
Supplementary Figure 8
Schematic of C. hominis chromosome VIII. (PDF 212 kb)
Supplementary Figure 9
Repeat distribution in C. hominis chromosomes. (PDF 66 kb)
Supplementary Figure 10
Gene ontology of C. hominis, P. falciparum, and Saccharomyces cerevisiae. (PDF 41 kb)
Supplementary Figure 11
Frequencies of polymorphisms in coding and non-coding sequences of C. hominis and C. parvum. (PDF 56 kb)
Supplementary Table 1A
Gene identification and putative function for C. hominis and C. parvum. C. hominis genome summary on individual chromosomes. (DOC 135 kb)
Supplementary Table 1B
Gene identification and putative function for C. hominis and C. parvum. (XLS 2622 kb)
Supplementary Table 2
C. hominis contigs and gaps aligned with C. parvum chromosomes. (XLS 226 kb)
Supplementary Table 3
Octamer repeat distributions. (DOC 45 kb)
Supplementary Table 4
Transfer RNAs and codon usage. (DOC 43 kb)
Supplementary Table 5
Metabolic pathways present in C. hominis, C. parvum, and P. falciparum. (XLS 139 kb)
Supplementary Table 6
Metabolic pathway overview of C. hominis and P. falciparum. (DOC 67 kb)
Supplementary Table 7
Genes associated with signal transduction and calcium regulation in C. hominis, C. parvum and P. falciparum. (XLS 17 kb)
Supplementary Table 8
Genes related to iron–sulphur complexes and mitochondria present in C. hominis, C. parvum and P. falciparum. (XLS 19 kb)
Supplementary Table 9
Organelle (non-mitochondrial) related proteins in C. hominis. (XLS 21 kb)
Supplementary Table 10
Potential drug targets in C. hominis. (DOC 70 kb)
Supplementary Table 11
Distribution of insertions and deletions in the C. hominis and C. parvum genomes. (XLS 1346 kb)
Supplementary Data 1
Contig accession numbers deposited in GenBank. (TXT 20 kb)
Supplementary Data 2
Gene accession numbers deposited in GenBank and KEGG. (TXT 49 kb)
Supplementary Data 3
rRNA accession numbers deposited in GenBank. (TXT 0 kb)
Supplementary Data 4
tRNA accession numbers deposited in GenBank. (TXT 0 kb)
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/), which permits distribution, and reproduction in any medium, provided the original author and source are credited. This licence does not permit commercial exploitation, and derivative works must be licensed under the same or similar licence.
About this article
Cite this article
Xu, P., Widmer, G., Wang, Y. et al. The genome of Cryptosporidium hominis. Nature 431, 1107–1112 (2004). https://doi.org/10.1038/nature02977
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02977
This article is cited by
-
Multiple pathways for glucose phosphate transport and utilization support growth of Cryptosporidium parvum
Nature Communications (2024)
-
Enrichment and proteomic identification of Cryptosporidium parvum oocyst wall
Parasites & Vectors (2022)
-
Diagnosis and control of cryptosporidiosis in farm animals
Journal of Parasitic Diseases (2022)
-
An update on Cryptosporidium biology and therapeutic avenues
Journal of Parasitic Diseases (2022)
-
Gregarine single-cell transcriptomics reveals differential mitochondrial remodeling and adaptation in apicomplexans
BMC Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.