Abstract
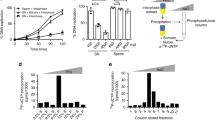
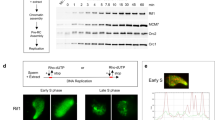
It is widely believed that DNA replication in multicellular animals (metazoa) begins at specific origins to which a pre-replicative complex (pre-RC) binds1. Nevertheless, a consensus sequence for origins has yet to be identified in metazoa. Origin identity can change during development, suggesting that there are epigenetic influences. A notable example of developmental specificity occurs in Drosophila, where somatic follicle cells of the ovary transition from genomic replication to exclusive re-replication at origins that control amplification of the eggshell (chorion) protein genes2. Here we show that chromatin acetylation is critical for this developmental transition in origin specificity. We find that histones at the active origins are hyperacetylated, coincident with binding of the origin recognition complex (ORC). Mutation of the histone deacetylase (HDAC) Rpd3 induced genome-wide hyperacetylation, genomic replication and a redistribution of the origin-binding protein ORC2 in amplification-stage cells, independent of effects on transcription. Tethering Rpd3 or Polycomb proteins to the origin decreased its activity, whereas tethering the Chameau acetyltransferase increased origin activity. These results suggest that nucleosome acetylation and other epigenetic changes are important modulators of origin activity in metazoa.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Bell, S. P. & Dutta, A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374 (2002)
Calvi, B. R., Lilly, M. A. & Spradling, A. C. Cell cycle control of chorion gene amplification. Genes Dev. 12, 734–744 (1998)
Claycomb, J. M., Benasutti, M., Bosco, G., Fenger, D. D. & Orr-Weaver, T. L. Gene amplification as a developmental strategy: isolation of two developmental amplicons in Drosophila. Dev. Cell 6, 145–155 (2004)
Spradling, A. & Mahowald, A. Amplification of genes for chorion proteins during oogenesis in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 77, 1096–1100 (1980)
Calvi, B. R. & Spradling, A. C. Chorion gene amplification in Drosophila: A model for metazoan origins of DNA replication and S-phase control. Methods 18, 407–417 (1999)
Calvi, B. R. & Spradling, A. C. The nuclear location and chromatin organization of active chorion amplification origins. Chromosoma 110, 159–172 (2001)
Royzman, I., Austin, R. J., Bosco, G., Bell, S. P. & Orr-Weaver, T. L. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes Dev. 13, 827–840 (1999)
Whittaker, A. J., Royzman, I. & Orr-Weaver, T. L. Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes Dev. 14, 1765–1776 (2000)
Bandura, J. L. & Calvi, B. R. Duplication of the genome in normal and cancer cell cycles. Cancer Biol. and Ther. 1, 8–13 (2002)
Austin, R. J., Orr-Weaver, T. L. & Bell, S. P. Drosophila ORC specifically binds to ACE3, an origin of DNA replication control element. Genes Dev. 13, 2639–2649 (1999)
de Cicco, D. & Spradling, A. Localization of a cis-acting element responsible for the developmentally regulated amplification of Drosophila chorion genes. Cell 38, 45–54 (1984)
Lu, L., Zhang, H. & Tower, J. Functionally distinct, sequence-specific replicator and origin elements are required for Drosophila chorion gene amplification. Genes Dev. 15, 134–146 (2001)
Claycomb, J. M., MacAlpine, D. M., Evans, J. G., Bell, S. P. & Orr-Weaver, T. L. Visualization of replication initiation and elongation in Drosophila. J. Cell Biol. 159, 225–236 (2002)
Vogelauer, M., Rubbi, L., Lucas, I., Brewer, B. J. & Grunstein, M. Histone acetylation regulates the time of replication origin firing. Mol. Cell 10, 1223–1233 (2002)
Mottus, R., Sobel, R. E. & Grigliatti, T. A. Mutational analysis of a histone deacetylase in Drosophila melanogaster: missense mutations suppress gene silencing associated with position effect variegation. Genetics 154, 657–668 (2000)
Bosco, G., Du, W. & Orr-Weaver, T. L. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nature Cell Biol. 3, 289–295 (2001)
Schwed, G., May, N., Pechersky, Y. & Calvi, B. R. Drosophila minichromosome maintenance 6 is required for chorion gene amplification and genomic replication. Mol. Biol. Cell 13, 607–620 (2002)
Lilly, M. & Spradling, A. The Drosophila endocycle is controlled by cyclin E and lacks a checkpoint ensuring S-phase completion. Genes Dev. 10, 2514–2526 (1996)
Cayirlioglu, P., Ward, W. O., Silver Key, S. C. & Duronio, R. J. Transcriptional repressor functions of Drosophila E2F1 and E2F2 cooperate to inhibit genomic DNA synthesis in ovarian follicle cells. Mol. Cell. Biol 23, 2123–2134 (2003)
Kadosh, D. & Struhl, K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12, 797–805 (1998)
Iizuka, M. & Stillman, B. Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. J. Biol. Chem. 274, 23027–23034 (1999)
Grienenberger, A. et al. The MYST domain acetyltransferase Chameau functions in epigenetic mechanisms of transcriptional repression. Curr. Biol. 12, 762–766 (2002)
Burke, T. W., Cook, J. G., Asano, M. & Nevins, J. R. Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J. Biol. Chem. 276, 15397–15408 (2001)
Simon, J. A. & Tamkun, J. W. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr. Opin. Genet. Dev. 12, 210–218 (2002)
Takei, Y. et al. MCM3AP, a novel acetyltransferase that acetylates replication protein MCM3. EMBO Rep. 2, 119–123 (2001)
Luo, R. X., Postigo, A. A. & Dean, D. C. Rb interacts with histone deacetylase to repress transcription. Cell 92, 463–473 (1998)
Kennedy, B. K., Barbie, D. A., Classon, M., Dyson, N. & Harlow, E. Nuclear organization of DNA replication in primary mammalian cells. Genes Dev. 14, 2855–2868 (2000)
Avni, D. et al. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol. Cell 12, 735–746 (2003)
Beall, E. L. et al. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420, 833–837 (2002)
Acknowledgements
We thank J. Simon and M. O'Connor for FRT Rpd3 flies; D. Buechle for hsp70:Gal4:Pc flies; M. Botchan, L. Beall, T. Orr-Weaver and S. Bell for ORC2 and Dup antibodies; D. Fyodorov and J. Kadonaga for Rpd3 antibody. We thank J. Claycomb for advice about QPCR. Thanks to N. May and M. Thomer for help with injections and flies. We are indebted to J. Bandura, A. K. Bielinsky, and M. Lilly for comments on the manuscript. This work was supported by a PHS grant to B.R.C.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Information
Includes supplementary methods and references (DOC 40 kb)
Supplementary Figure Legends
Figure S1 Developmental time course of BrdU and AcH4 labeling; Figure S2 Quantification of AcH4, ORC2, and TOTO-3 DNA label at the 3rd chorion locus; Figure S3 Acetylated foci are evident despite reduced chorion copy number in the mutant MCM6K1214; Figure S4 Antibodies for other acetylated forms of histone label chorion foci; Figure S5 Antibody labeling of Rpd3 mutant clones with Rpd3 antibody indicates they have reduced Rpd3 protein; Figure S6 Antibodies raised against other modifications of histone H4 indicate Rpd3 clones have widespread hyperacetylation; Figure S7 Quantification of ectopic BrdU in Rpd3 mutant clones; Figure S8 Quantification of ectopic ORC2 in Rpd3 mutant clones. (DOC 30 kb)
Rights and permissions
About this article
Cite this article
Aggarwal, B., Calvi, B. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430, 372–376 (2004). https://doi.org/10.1038/nature02694
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature02694
This article is cited by
-
Rpd3/CoRest-mediated activity-dependent transcription regulates the flexibility in memory updating in Drosophila
Nature Communications (2021)
-
Su(Hw) primes 66D and 7F Drosophila chorion genes loci for amplification through chromatin decondensation
Scientific Reports (2021)
-
Anatomy and evolution of a DNA replication origin
Chromosoma (2021)
-
Order from clutter: selective interactions at mammalian replication origins
Nature Reviews Genetics (2017)
-
Widespread colocalization of the Drosophila histone acetyltransferase homolog MYST5 with DREF and insulator proteins at active genes
Chromosoma (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.