Abstract
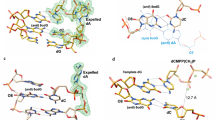
Ultraviolet light damages DNA by catalysing covalent bond formation between adjacent pyrimidines, generating cis–syn cyclobutane pyrimidine dimers (CPDs) as the most common lesion1. CPDs block DNA replication by high-fidelity DNA polymerases, but they can be efficiently bypassed by the Y-family DNA polymerase pol η2,3. Mutations in POLH encoding pol η are implicated in nearly 20% of xeroderma pigmentosum, a human disease characterized by extreme sensitivity to sunlight and predisposition to skin cancer4,5,6. Here we have determined two crystal structures of Dpo4, an archaeal pol η homologue, complexed with CPD-containing DNA, where the 3′ and 5′ thymine of the CPD separately serves as a templating base. The 3′ thymine of the CPD forms a Watson–Crick base pair with the incoming dideoxyATP, but the 5′ thymine forms a Hoogsteen base pair with the dideoxyATP in syn conformation. Dpo4 retains a similar tertiary structure, but each unusual DNA structure is individually fitted into the active site for catalysis. A model of the pol η–CPD complex built from the crystal structures of Saccharomyces cerevisiae apo-pol η and the Dpo4–CPD complex suggests unique features that allow pol η to efficiently bypass CPDs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Ravanat, J. L., Douki, T. & Cadet, J. Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B 63, 88–102 (2001)
Masutani, C., Kusumoto, R., Iwai, S. & Hanaoka, F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 19, 3100–3109 (2000)
Johnson, R. E., Washington, M. T., Prakash, S. & Prakash, L. Fidelity of human DNA polymerase η. J. Biol. Chem. 275, 7447–7450 (2000)
Lehmann, A. R. Replication of damaged DNA in mammalian cells: new solutions to an old problem. Mutat. Res. 509, 23–34 (2002)
Masutani, C. et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399, 700–704 (1999)
Johnson, R. E., Kondratick, C. M., Prakash, S. & Prakash, L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285, 263–265 (1999)
Thoma, F. Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J. 18, 6585–6598 (1999)
Hoeijmakers, J. H. Genome maintenance mechanisms for preventing cancer. Nature 411, 366–374 (2001)
Ohmori, H. et al. The Y-family of DNA polymerases. Mol. Cell 8, 7–8 (2001)
Yang, W. Damage repair DNA polymerases Y. Curr. Opin. Struct. Biol. 13, 23–30 (2003)
Ling, H., Boudsoçq, F., Woodgate, R. & Yang, W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107, 91–102 (2001)
Boudsocq, F., Iwai, S., Hanaoka, F. & Woodgate, R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DNA polymerase with properties akin to eukaryotic Pol η. Nucleic Acids Res. 29, 4607–4616 (2001)
Patel, P. H., Suzuki, M., Adman, E., Shinkai, A. & Loeb, L. A. Prokaryotic DNA polymerase I: evolution, structure, and “base flipping” mechanism for nucleotide selection. J. Mol. Biol. 308, 823–837 (2001)
Glick, E., Vigna, K. L. & Loeb, L. A. Mutations in human DNA polymerase η motif II alter bypass of DNA lesions. EMBO J. 20, 7303–7312 (2001)
Park, J. Y. & Choi, B. S. NMR investigation of echinomycin binding to d(ACGTTAACGT)2: Hoogsteen versus Watson-Crick A.T base pairing between echinomycin binding sites. J. Biochem. (Tokyo) 118, 989–995 (1995)
Aishima, J. et al. A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 30, 5244–5252 (2002)
Patikoglou, G. A. et al. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 13, 3217–3230 (1999)
Bunting, K. A. et al. Crystal structure of the Escherichia coli dcm very-short-patch DNA repair endonuclease bound to its reaction product-site in a DNA superhelix. Nucleic Acids Res. 31, 1633–1639 (2003)
Park, H. et al. Crystal structure of a DNA decamer containing a cis–syn thymine dimer. Proc. Natl Acad. Sci. USA 99, 15965–15970 (2002)
Zhang, H. & Siede, W. UV-induced T → C transition at a TT photoproduct site is dependent on Saccharomyces cerevisiae polymerase η in vivo. Nucleic Acids Res. 30, 1262–1267 (2002)
Szekeres, E. S. Jr, Woodgate, R. & Lawrence, C. W. Substitution of mucAB or rumAB for umuDC alters the relative frequencies of the two classes of mutations induced by a site-specific T-T cyclobutane dimer and the efficiency of translesion DNA synthesis. J. Bacteriol. 178, 2559–2563 (1996)
Trincao, J. et al. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell 8, 417–426 (2001)
Washington, M. T., Prakash, L., Prakash, S. & Yeast, D. N. A. polymerase η utilizes an induced-fit mechanism of nucleotide incorporation. Cell 107, 917–927 (2001)
Zhou, B. L., Pata, J. D. & Steitz, T. A. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell 8, 427–437 (2001)
Silvian, L. F., Toth, E. A., Pham, P., Goodman, M. F. & Ellenberger, T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nature Struct. Biol. 8, 984–989 (2001)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Brünger, A. T. et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Jones, T. A., Zou, J.-Y. & Cowan, S. W. Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Vassylyev, D. G. et al. Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell 83, 773–782 (1995)
Cohen, G. E. ALIGN: a program to superimpose protein coordinates. J. Appl. Crystallogr. 30, 1160–1161 (1997)
Acknowledgements
We thank R. Craigie and D. Leahy for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
41586_2003_BFnature01919_MOESM1_ESM.gif
Supplementary Animation 1: Structural comparison of TT-1, TT-2 and the Type I Dpo4 and undamaged DNA complex 11. The three structures are shown sequentially after superposition of the palm, finger and thumb domains of Dpo4. The protein domains are colour coded as in Fig. 3a, and the DNA is shown in a blue stick model. The incoming nucleotide is highlighted in yellow, and the thymine dimer in orange. The metal ions are shown as grey spheres. There is only one metal ion in the type I structure because the incoming ddATP was hydrolyzed to ddADP. (GIF 177 kb)
41586_2003_BFnature01919_MOESM2_ESM.gif
Supplementary Animation 2: Modeling of Pol eta complexed with a CPD. The crystal structure of Pol eta apo-protein presented in a multi-colour ribbon diagram and the TT-1 structure in grey-blue are shown together after superimposing the palm domains of Pol eta and Dpo4. For Pol eta to interact with the DNA substrate as the domains of Dpo4 do in TT-1, the finger, thumb and little finger domains of Pol eta have to rotate 16°, 16° and 45°, respectively, toward the DNA as shown in the next frame. (GIF 115 kb)
Rights and permissions
About this article
Cite this article
Ling, H., Boudsocq, F., Plosky, B. et al. Replication of a cis–syn thymine dimer at atomic resolution. Nature 424, 1083–1087 (2003). https://doi.org/10.1038/nature01919
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature01919
This article is cited by
-
Backbone assignment of the binary complex of the full length Sulfolobus solfataricus DNA polymerase IV and DNA
Biomolecular NMR Assignments (2017)
-
Molecular architecture of the Ub-PCNA/Pol η complex bound to DNA
Scientific Reports (2015)
-
A nucleotide binding rectification Brownian ratchet model for translocation of Y-family DNA polymerases
Theoretical Biology and Medical Modelling (2011)
-
Structural insight into dynamic bypass of the major cisplatin-DNA adduct by Y-family polymerase Dpo4
The EMBO Journal (2010)
-
Replication through an abasic DNA lesion: structural basis for adenine selectivity
The EMBO Journal (2010)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.