Abstract
The Trail Making Test (TMT) is a widely used test of executive function and has been thought to be strongly associated with general cognitive function. We examined the genetic architecture of the TMT and its shared genetic aetiology with other tests of cognitive function in 23 821 participants from UK Biobank. The single-nucleotide polymorphism-based heritability estimates for trail-making measures were 7.9% (part A), 22.4% (part B) and 17.6% (part B−part A). Significant genetic correlations were identified between trail-making measures and verbal-numerical reasoning (r g>0.6), general cognitive function (r g>0.6), processing speed (r g>0.7) and memory (r g>0.3). Polygenic profile analysis indicated considerable shared genetic aetiology between trail making, general cognitive function, processing speed and memory (standardized β between 0.03 and 0.08). These results suggest that trail making is both phenotypically and genetically strongly associated with general cognitive function and processing speed.
Similar content being viewed by others
Introduction
The Trail Making Test (TMT) is widely used in both research and clinical settings as a test of some aspects of executive function.1,2,3 The TMT is usually given as two parts, from which three measures are derived. In TMT Part A (TMT A), participants are required to connect an array of numbers in ascending order, by drawing a continuous line (trail) between them as quickly and accurately as possible. TMT Part B (TMT B) requires participants to connect an array of both numbers and letters in alternating ascending order (1, A, 2, B, 3, C and so on) with the same emphasis on speed and accuracy (Supplementary Figure 1). Subtracting TMT A completion time from that of TMT B (TMT B−A) is thought to allow the relative contributions of visual search and psychomotor speed to be parsed from the more complex executive functions (such as cognitive flexibility) required to alternate between numbers and letters.4,5,6,7,8
TMT performance has been ascribed to a number of cognitive processes, ‘including attention, visual search and scanning, sequencing and shifting, psychomotor speed, abstraction, flexibility, ability to execute and modify a plan of action, and ability to maintain two trains of thought simultaneously’.9 It is considered a useful tool in research and clinical practice due to the sensitivity of the task (particularly TMT B and B−A) to frontal lobe damage (in some, but not other studies10) and dementia.11,12,13 There are declines in both TMT A and B performance in ageing.10,14,15,16,17,18,19,20 There is also evidence for performance deficits on TMT B in mood disorders21 and in patients with schizophrenia and their relatives18,22,23,24,25,26,27
Family-based and twin-based studies have provided evidence for a genetic contribution to individual differences in trail making, estimating the heritability for trail making part A between 0.23 and 0.38, and between 0.39 and 0.65 for trail making part B.28,29,30 A recent genome-wide association study (GWAS) of trail making part A and part B in a sample of around 6000 individuals did not find any genome-wide significant hits;31 however, GWAS of other cognitive phenotypes have demonstrated that much larger sample sizes are required to reliably identify significant genetic loci.32,33 Trail making is thought to have genetic influences that are shared with other cognitive abilities, with a twin-based genetic correlation of 0.48 reported between trail making, measured as the ratio between trail making part A and trail making part B, and general cognitive function, and 0.52 with working memory.34
In addition to a relatively poor understanding of the molecular genetic underpinnings of TMT, its cognitive and psychometric architecture merits further research. Specifically, it is unclear whether the cognitive abilities required for TMT B performance are distinct from other cognitive domains, because TMT A and B scores correlate with measures of general cognitive function and processing speed (correlation coefficient estimates range from ~0.3 to ~0.7 (refs9,20)).30,35,36,37,38 Evidence from cognitive-ageing studies further suggests a strong overlap between TMT performance and other cognitive domains, because age-related decline in processing speed and working memory account for much of the age effects on TMT B.9,20,39,40 However, reliably identifying the cognitive processes that underpin cognitive test performance using phenotypic correlational analyses alone is sub-optimal, for example, refs41,42,43. Rather, the interpretation of relationships between TMT performance and other cognitive abilities such as general cognitive function, may be enhanced by considering and comparing their respective shared and unshared causes, including their genetic architecture. By examining the shared genetic architecture of TMT and other measures of cognitive ability, the current study might aid a better understanding of potential biological pathways involved in general and specific cognitive abilities.
The aim of the present study is to (1) add to the understanding of the genetic architecture of trail making and (2) to explore the overlap between genetic architecture of TMT performance and other cognitive abilities, including general cognitive function. Although several studies have examined the genetic architecture of the TMT and its overlap with other cognitive abilities, they had relatively low power. The largest TMT GWAS (N~6000) to date used a variety of assessments across multiple cohorts across multiple countries, potentially leading to heterogeneity in both sample composition and in the cognitive measures.31 The current study design, using >23 000 UK Biobank participants, mitigates such confounds: the almost fourfold increase in sample size yields greater statistical power, and the single sample reduces heterogeneity in genetic testing and in phenotype, because all participants had white British ancestry and took the same TMT test with the same instructions. In addition, we add to this significantly larger GWAS than has previously been conducted with (1) estimates of the single-nucleotide polymorphism (SNP)-based heritability of TMT performance and (2) an examination of genetic overlap among the different TMT test measures and with other cognitive functions, using polygenic profile analysis and genetic correlations in independent samples.
Materials and methods
Participants
UK Biobank is a large resource which aims to identify determinants of human diseases in middle aged and older individuals (http://www.ukbiobank.ac.uk).44 A total of 502 655 community-dwelling individuals aged between 37 and 73 years were recruited in the United Kingdom between 2006 and 2010. Baseline assessment included cognitive testing, personality self-report, and physical and mental health measures. For the present study, genome-wide genotyping data were available for 112 151 participants (58 914 females and 53 237 males) after quality control (see below). They had a mean (s.d.) age of 56.9 (7.9) years (range 40 to 73 years). UK Biobank received ethical approval from the Research Ethics Committee (REC reference for UK Biobank is 11/NW/0382). This study was completed under UK Biobank application 10279. Figure 1 shows the study flow for the present study.
Measures
Trail making test
The TMT A and TMT B were introduced at a follow-up testing wave in UK Biobank, between 2014 and 2015. For TMT A, participants were instructed to connect numbers (1–25) consecutively (which were quasi-randomly distributed on the touchscreen) as quickly as possible in ascending order by selecting the next number. TMT B is similar, but letters (A–L) and numbers (1–13) had to be selected in alternating ascending order, for example, 1A 2 B 3 C and so on. The intervals between touching two points was timed in seconds using a Javascript timer. The total time (in seconds) to complete the TMT (part A or B) was derived by summing the interval values between two points. Nine out of 23 821 individuals who scored >250 s for TMT B were excluded. Owing to positively skewed distributions, both TMT A and TMT B scores were log-transformed prior to further analyses. The difference between the raw scores for TMT A and TMT B was computed as TMT B minus TMT A (TMT B−A). Fifty-two out of 23 821 individuals with scores <−50 or >150 were removed from TMT B−A. After exclusions, 23 822 individuals with genetic data completed TMT A, 23 812 individuals with genetic data completed TMT B and 23 769 individuals had complete information for TMT B−A. The three trail making measures have been scored such that a higher score indicates better performance. This study also used the verbal-numerical reasoning (VNR) test from UK Biobank (VNR, N=36 035), which consisted of a 13-item questionnaire assessing verbal and arithmetical deduction (Cronbach α reliability=0.62) as described by Hagenaars et al. 45
Genotyping and quality control
The interim release of UK Biobank included genotype data for 152 729 individuals, of whom 49 979 were genotyped using the UK BiLEVE array and 102 750 using the UK Biobank axiom array. These arrays have over 95% content in common. Details of the array design, genotyping procedures and quality control details have been published elsewhere.45,46 UK Biobank released an imputed dataset as part of the interim data release. More details can be found at the following URL: http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=157020. A minor allele frequency cutoff of 1% was used for all autosomal variants, as well as an imputation quality score above 0.3 (N~9.7M SNPs).
GWAS analysis
Genotype–phenotype association analyses were conducted using SNPTEST v2.5.1.47 The ‘frequentist 1’ option was used to specify an additive model and genotype dosages were analysed to account for genotype uncertainty. All phenotypes were adjusted for the following covariates before analysis; age, gender, assessment centre, genotyping array and batch, and the first 10 genetic principal components for population stratification. The GWAS of VNR has been performed previously.32
SNP-based heritability and genetic correlations
Univariate GCTA-GREML48 analysis was performed to estimate the proportion of variance explained by all genotyped common SNPs for TMT A, TMT B and TMT B−A. To include only unrelated individuals, a relatedness cutoff of 0.025 was used in the generation of the genetic relationship matrix. Given the likely difference in reliability between the raw (TMT A and TMT B) and difference (TMT B−A) scores,49 we undertook a supplementary analysis to compare reliability-weighted heritability estimates (see Supplementary Methods) to provide a clearer comparison of h2 differences between elements of the TMT. Bivariate GCTA-GREML48 and linkage disequilibrium (LD) score regression50 were used to derive genetic correlations between TMT measures and VNR in UK Biobank.32 LD score regression was also used to estimate genetic correlations between trail making measures in UK Biobank and trail making part A, trail making part B, general cognitive function, processing speed and memory from the CHARGE consortium (participants aged 45 years or older) meta-analyses of these cognitive phenotypes.31,33,51
Gene-based association analysis
MAGMA52 was used to derive gene-based associations using the summary results of the three GWAS for trail making. Genes (18 354) were analysed after the SNPs were assigned a gene based on their position using the NCBI 37.3 build, without additional boundaries placed around the genes. To account for LD, the European 1000 Genomes data panel (phase 1, release 3) was used as a reference. A Bonferroni correction was used to control for 18 345 tests (α=0.05/18 345; P<2.73 × 10−6). The gene-based associations for trail making were compared with gene-based associations for VNR and with the gene-based associations for trail making, general cognitive function, processing speed and memory from the CHARGE consortium, based on the GWAS summary results.31,33,51 The CHARGE summary results were converted from HapMap2 to 1000G format, to ensure the maximum overlap between the two samples. This was achieved using the LiftOver programme, which converts coordinate ranges between genome assemblies.
Partitioned heritability
Partitioned heritability analyses were performed on the trail making SNP-based association results to determine if SNPs group together according to a specific biological function or role and thereby making an enriched contribution to the total proportion of heritability of the trail making phenotypes. These analyses were performed using the data processing pipeline as suggested by Finucane et al. 53
Polygenic profile analyses
Polygenic profile analyses were performed to predict TMT performance into UK Biobank and to predict cognitive function scores in two independent cohorts, Generation Scotland’s Scottish Family Health Study (GS) and the Lothian Birth Cohort of 1936 (LBC1936). Prediction from the CHARGE consortium meta-analysis of trail making, general cognitive function, processing speed and memory into UK Biobank was performed to test the extent to which individual differences in TMTs in UK Biobank could be predicted by the polygenic architecture of these four traits.
Polygenic prediction into UK Biobank
The UK Biobank genotyping data were recoded from numeric (1,2) allele coding to standard ACGT coding using a bespoke programme developed by one of the present authors (DCL).45 Polygenic profile scores were created for trail making part A, trail making part B, general cognitive function, processing speed and memory based on the results from the CHARGE consortium meta-analysis in all genotyped participants using PRSice.54 SNPs with a minor allele frequency<0.01 were removed before creating the scores. Clumping was used to obtain SNPs in LD with an r 2<0.25 within a 250 kb window. Five polygenic profiles were created for each of the three phenotypes according to the significance of the association with the trail making phenotype, at P-value thresholds of 0.01, 0.05, 0.1, 0.5 and 1 (all LD pruned SNPs). Regression models were used to examine the association between the polygenic profiles and TMT A, TMT B and TMT B−A phenotype scores in UK Biobank, adjusting for age at measurement, sex, genotyping batch and array, assessment centre and the first 10 genetic principal components for population stratification. All associations were corrected for multiple testing using the false discovery rate method.55
Polygenic prediction into GS and LBC1936
Polygenic profile scores were created in PRSice,54 using the UK Biobank trail making SNP-based association results, for genotyped participants of GS (n=19 994, mean (s.d.) age=47.18 (15.10) years) and LBC1936 (n=1005, mean (s.d.) age of 72.55 (0.7) years for all tests, except trail making part B, which was tested at a mean (s.d.) age of 76.30 (0.68) years). Individuals were removed from GS if they had contributed to both UK Biobank and GS (n=174). Polygenic profile scores were created based on the significance of the association in UK Biobank with the trail making phenotype, at P-value thresholds of 0.01, 0.05, 0.1, 0.5 and 1 (all SNPs). Linear regression models were created to test the association between the polygenic profiles for trail making and the target phenotypes in: GS (Wechsler digit-symbol substitution,56 phonemic verbal fluency,1 Wechsler logical memory,57 the Mill Hill vocabulary test,58 a fluid cognitive function component and a general cognitive function component) and in LBC1936 (trail making part B, a fluid cognitive function component, vocabulary, memory, processing speed, change in fluid cognitive function between age 11 years and age 70 years, Moray House Test score at age 11 years and Moray House Test score at age 70 years). Further details about the cognitive tests can be found in the Supplementary Materials. All models were adjusted for age, sex and the first five (GS) or four (LBC1936) principal components for population stratification. The GS models were also adjusted for family structure by fitting a univariate linear mixed model, which estimates the genetic and environmental variance, using the ASReml programme.59 All associations were corrected for multiple testing using the false discovery rate method.55
Meta-analysis
Inverse variance-weighted meta-analysis of UK Biobank trail making and CHARGE trail making GWAS was performed using the METAL package (http://www.sph.umich.edu/csg/abecasis/Metal). The meta-analysis was restricted to SNPs that were available in both samples (N SNPs TMT A=2 332 746; N SNPs TMT B=2 466 810) and the samples did not include any overlapping individuals. The total sample in the meta-analysis consisted of 29 251 individuals for TMT A and 30 022 for TMT B.
Code availability
The code used to run the analysis is available from the authors upon request.
Results
Phenotypic correlations
A total of 23 822 individuals with genetic data completed TMT A, 23 812 individuals with genetic data completed TMT B and 23 769 individuals with genetic data had complete information for TMT B−A. Table 1 shows the phenotypic correlations between the TMT phenotypes, as well as with VNR in UK Biobank. Correlations indicated that individuals who took more time to complete the TMTs had lower performance on the VNR test. The strongest correlation between VNR and TMT was found for TMT B (0.36). Supplementary Figure 2 shows the age and sex distribution of the different trail making measures.
Genome-wide association study
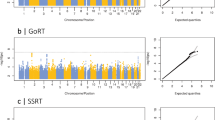
The results of the GWAS analyses are presented in Figure 2; for each trail making phenotype, a Manhattan and a Q–Q plot are shown.
Manhattan and Q–Q plot of P-values of the single-nucleotide polymorphism (SNP)-based association analysis of Trail Making Part A, Trail Making Part B and Trail Making Part B−Part A. The red line in the Manhattan plots indicates the threshold for genome-wide significance (P<5 × 10−8); the grey line indicates the threshold for suggestive significance (P<1 × 10−5).
TMT part A
For TMT A, no SNPs reached genome-wide significance. Gene-based analyses identified three genes that were significantly associated with TMT A (Table 2 and Supplementary Table 1a); CRNKL1, a protein necessary for pre-mRNA splicing on chromosome 20;60 caspase 5 (CASP5), which has a role in the execution phase of cell apoptosis on chromosome 11;61 and NAA20, a component of the N-acetyltransferase complex B on chromosome 20.62 All three of these genes were also nominally significant (P<0.05) in the GWAS of TMT B and CASP5 was also nominally significant in the analyses for CHARGE general cognitive function and processing speed (Table 2).
The proportion of variance in TMT A explained by all common genetic variants using GCTA-GREML was 0.079 (SE 0.024).
TMT part B
One SNP exhibited genome-wide significance for TMT B, on chromosome 1 (rs34804445, P=4.18 × 10−8, MAF=0.0253). This SNP was not located within a gene. Gene-based analyses identified one gene (PUM1 on chromosome 1) associated with TMT B (Table 2 and Supplementary Table 1b); this gene regulates ATAXIN1 and is associated with neurodegeneration.63 This gene was also nominally significant (P<0.05) in the gene-based analysis for the other trail making variables measured in UK Biobank. Results for this gene were not available for the CHARGE consortium phenotypes (Table 2).
The proportion of variance in TMT B explained by all common genetic variants using GCTA-GREML was 0.224 (s.e. 0.026).
TMT part B−A
No SNPs reached genome-wide significance for TMT B−A. Gene-based analyses did not identify any significant associations for TMT B−A (Supplementary Table 1c).
The proportion of variance in TMT B−A explained by all common genetic variants using GCTA-GREML was 0.176 (s.e. 0.025). Heritability estimates and s.e. of all three TMT measures, weighted according to estimates of their reliabilities, are reported in Supplementary Table 2. They indicate that the ostensible difference in h2 between TMT B and TMT B−A could possibly be attributed to differences in reliability.
Partitioned heritability
Significant enrichment was found for TMT B in evolutionary conserved regions with a 500 bp boundary, where 33% of the SNPs accounted for 95% of the heritability (enrichment metric=2.85, s.e.=0.44, P=2.42 × 10−5). TMT A and TMT B−A were unsuitable for this analysis, due to low heritability Z-scores, which were 2.6 and 5.4, respectively.53
Genetic correlations
The results of the bivariate GCTA-GREML and LD score regression analyses within UK Biobank are shown in Table 3. The LD score regression analyses with the cognitive phenotypes from the CHARGE consortium are shown in Table 4. Strong positive genetic correlations, using GCTA-GREML, were observed between all three measures derived from the TMT in UK Biobank (r g between 0.64 and 0.96). Large genetic correlations were found between the three TMTs and VNR (r g between 0.59 and 0.64). Similar results were found when calculating the genetic correlations using LD score regression (Table 3). Positive genetic correlations were observed between trail making in UK Biobank and the following GWAS meta-analyses from the CHARGE consortium: general cognitive function (r g between 0.61 and 0.70), processing speed (r g between 0.69 and 0.76) and memory (r g between 0.29 and 0.35) (Table 4). The confidence interval for the associations between TMT and general cognitive function, and between TMT and processing speed did not overlap with the confidence interval for the association between TMT and memory.
Polygenic prediction
Polygenic profiles based on the TMT A summary results from the CHARGE consortium significantly predicted all three TMTs in UK Biobank (β between 0.016 and 0.029, Table 5). Polygenic profiles based on the TMT B summary results from the CHARGE consortium significantly predicted all three TMTs in UK Biobank (β between 0.024 and 0.036, Table 5). The strongest associations for the other cognitive test polygenic profiles (general cognitive function, processing speed and memory) were found for general cognitive function polygenic profiles predicting TMT B in UK Biobank, explaining 0.67% of the variance. The full results including all thresholds can be found in Supplementary Table 3a.
The GWAS results for the three trail making phenotypes in UK Biobank were used to create polygenic profile scores in two independent cohorts: GS and the LBC1936 (Figure 3 and Supplementary Tables 3b and 3c). The polygenic profiles for TMT A significantly predicted digit-symbol substitution, verbal fluency, Mill Hill vocabulary, fluid cognitive function and general cognitive function in GS (β between 0.018 and 0.039). In the LBC1936, the polygenic profiles for TMT A significantly predicted trail making part B (β=0.102), fluid cognitive function, memory and change in fluid cognitive function (β between 0.079 and 0.094). Significant predictions were observed across almost all thresholds for TMT B and TMT B−A for the cognitive phenotypes measured in GS. In LBC1936, polygenic profiles for TMT B and TMT B−A were both significantly associated with fluid cognitive function, processing speed, cognitive function at age 11 years and cognitive function at age 70 years. TMT B was also significantly associated with trail making part B, memory and change in fluid cognitive function between age 11 years and age 70 years. The strongest association in GS was found between the polygenic profile for TMT B and Wechsler digit symbol substitution; this association explained 0.53% of the variance, at an SNP inclusion threshold of all SNPs from the GWAS. The largest proportion of variance explained in LBC1936 was 1.75% for trail making part B using the TMT B polygenic score with an SNP inclusion threshold of all SNPs from the GWAS. The complete results can be found in Supplementary Tables 3b and c.
Heat map of associations between the polygenic profile scores for trail making in UK Biobank and cognitive function in Generation Scotland (GS) and the Lothian Birth Cohort 1936 (LBC1936). *False discovery rate (FDR)-corrected significant associations (FDR corrected P-value <=0.029 (GS) or 0.025 (LBC1936)). Further information can be found in Supplementary Tables 2b and c. Age 11 MHT score, age 11 Moray House Test score; Age 70 MHT score, age 70 Moray House Test score; Vocabulary (MHV), Mill Hill Vocabulary test; Vocabulary (NART), National Adult Reading Test; LBC1936, Lothian Birth Cohort 1936; TMT A, Trail Making Test part A; TMT B, Trail Making Test part B; TMT B−A, Trail Making Test part B−Trail Making Test part A; Stand. β, standardized β.
Meta-analysis
In the meta-analysis of the combined dataset (UK Biobank and CHARGE, combined N TMT A=29 251, combined N TMT B=30 022), no genome-wide significant SNPs were observed for TMT A or TMT B (Supplementary Figure 3). The one genome-wide significant hit identified in this study for TMT B was not available in CHARGE.
Discussion
This study finds one genome-wide significant variant associated with TMT part B performance in UK Biobank, and provides the first SNP-based heritability estimates for the three widely used TMT measures. We identified high genetic correlations between trail making, VNR, general cognitive function and processing speed, and somewhat lower genetic correlations with memory. Using only common SNPs to create a polygenic score for TMT B, we were able, at best, to predict ~2% of the variance in trail making part B in LBC1936. Taken together, these analyses point to a considerable degree of shared polygenic architecture for TMT performance with general cognitive function and processing speed measures, in particular. The results are consistent with the hypothesis that trail making is genetically and phenotypically similar to general fluid cognitive function.9,20,34
Univariate GCTA-GREML analyses suggested SNP-based heritability estimates for TMT A, TMT B and TMT B−A of 8%, 22% and 18%, respectively. These provide a first estimate of the contribution of common SNPs to the phenotype of trail making and suggest that common SNPs account for around half of the additive genetic variation for trail making, based on twin and family studies.30 SNP-based heritability studies for other complex traits also report similar differences between SNP-based and pedigree-based heritability.48,64 Twin and family studies estimate heritability based on all causal variants, both common and rare, whereas SNP-based studies estimate heritability only on genotyped SNPs in LD with causal variants. It is possible that causal variants have lower minor allele frequencies than the genotyped SNPs, leading to incomplete LD between unknown causal variants and genotyped SNPs.65
Previous studies of the genetic overlap between trail making and other cognitive abilities have shown genetic overlap between trail making and general cognitive function using a twin design.30,34 Our results add to this by using a molecular genetic design and showing shared genetic aetiology between trail making and verbal numerical reasoning in UK Biobank, as well as with general cognitive function, processing speed, and memory from the CHARGE consortium. The estimates of the genetic correlations within UK Biobank were similar for both GCTA-GREML and LDS regression, suggesting that these results are unlikely to constitute false positives.
We also used polygenic profile scores to estimate the genetic overlap between TMT and other cognitive abilities. First, we created polygenic profiles based on the CHARGE consortium summary GWAS data (including trail making, general cognitive function, processing speed and memory) and found that the polygenic profile for general cognitive function was the best predictor of all TMT scores in UK Biobank compared with either of the other polygenic profiles. The unequal sample sizes from which the polygenic profile scores were derived (TMT~6000; general cognitive function ~54 000; processing speed~32 000, memory~29 000) may have led to an underestimation of the ability of CHARGE TMT score to predict UK Biobank TMT test performance.
The genetic association between TMT and other cognitive measures could be due to an overall halo-effect of general cognitive ability. Although the current study does not allow us to test this empirically, we note that the respective polygenic associations between TMT and fluid cognitive ability in GS and LBC1936, as well as the genetic correlations between TMT and VNR, do point to this halo-effect. Future studies could expand these findings by removing the variance specific to general cognitive ability from the associations between TMT and other measures of cognitive ability.
Our combined meta-analysis of UK Biobank and CHARGE trail making did not yield any significant SNPs. This could be ascribed to the following factors. The SNPs for the CHARGE consortium are imputed to the HAPMAP 2 reference panel (N~2M SNPs), whereas the UK Biobank sample is imputed to a combination of the UK10 haplotype and the 1000G reference panel (N~17M SNPs). For TMT A 2 332 746 SNPs overlapped between UK Biobank and CHARGE and for TMT B 2 466 810 SNPs overlapped between the two samples. The SNP that reached genome-wide significance in the UK Biobank GWAS was not available in CHARGE.
In addition to those discussed above, our study has other limitations. There are currently no reliability data available for the TMT version administered in UK Biobank. Although the computer-based version was based upon standard administration protocols, the current absence of such data necessitates that these results be interpreted with appropriate caution. The measure of fluid cognitive function provided by UK Biobank (which we have called VNR) showed a relatively modest age-related trajectory, in contrast to the steeper and well-replicated age-related decline that would be expected for this construct66,67 (Supplementary Figure 4). This may partly explain the relatively modest correlation of VNR with TMT performance, when compared to those previously reported9 and may have also had a bearing on our analyses of genetic overlap within UK Biobank. In addition, the UK Biobank sample did not have sufficient breadth of contemporaneously-administered standardized/validated cognitive tests to be able to construct a robust measure of general cognitive function (such as in other large samples9,20,68). This limited our ability to perform a more detailed analysis of the phenotypic or genetic overlap between trail making and general cognitive function in UK Biobank itself.
This study has several strengths. It has the largest single sample size to date of a GWAS for trail making, offering greater statistical power while excluding bias caused by sample or phenotypic heterogeneity. Population stratification was minimized by only using individuals of white British ancestry. This study shows the first estimates of the heritability for TMT using molecular genetic data uses a comprehensive battery of techniques to examine genetic architecture of TMT and used the same TMT and administration protocol across the whole sample. The use of both GWAS summary data from the CHARGE consortium, and the prediction of the UK Biobank TMT GWAS into GS and LBC1936 cognitive phenotypes allowed a detailed examination of the shared genetic aetiology between TMT and cognitive phenotypes.
These strengths have enabled a detailed characterization of the shared genetic aetiology between performance on TMT and other cognitive abilities. Our results, spanning methodologies and cohorts, provide strong evidence for a shared genetic aetiology between TMT performance and general cognitive function and processing speed, which are themselves strongly phenotypically and genetically correlated. When the full genetic data from UK Biobank on half a million individuals becomes available, it would enable robust replication and extensions of the current findings. More detailed cognitive testing is planned for UK Biobank, and these data can be used in future studies to further examine the genetic overlap between TMT and other cognitive functions.
References
Lezak MD, Howieson DB, Loring DW. Neuropsychological Assessment. Oxford University Press: USA, 2004.
Reitan RM, Wolfson D. The Halstead-Reitan Neuropsychological Test Battery: Theory and Clinical Interpretation, vol. 4 Neuropsychology Press: Tucson, AZ, 1985.
Delis DC, Kaplan E, Kramer JH. Delis-Kaplan Executive Function System (D-KEFS). Psychological Corporation: San Antonio, TX, 2001.
Bowie CR, Harvey PD. Administration and interpretation of the Trail Making Test. Nat Protoc 2006; 1: 2277–2281.
Strauss E, Sherman EM, Spreen O. A Compendium of Neuropsychological Tests: Administration, Norms, and Commentary. American Chemical Society: New York, NY, 2006.
Arbuthnott K, Frank J. Trail making test, part B as a measure of executive control: validation using a set-switching paradigm. J Clin Exp Neuropsychol 2000; 22: 518–528.
Kortte KB, Horner MD, Windham WK. The trail making test, part B: cognitive flexibility or ability to maintain set? Appl Neuropsychol 2002; 9: 106–109.
Gläscher J, Adolphs R, Damasio H, Bechara A, Rudrauf D, Calamia M et al. Lesion mapping of cognitive control and value-based decision making in the prefrontal cortex. Proc Natl Acad Sci USA 2012; 109: 14681–14686.
Salthouse TA. What cognitive abilities are involved in trail-making performance? Intelligence 2011; 39: 222–232.
MacPherson SE, Della Sala S, Cox SR, Girardi A, Iveson MH. Handbook of Frontal Lobe Assessment. OUP Oxford, 2015.
Amieva H, Lafont S, Auriacombe S, Rainville C, Orgogozo J-M, Dartigues J-F et al. Analysis of error types in the Trail Making Test evidences an inhibitory deficit in dementia of the Alzheimer type. J Clin Exp Neuropsychol 1998; 20: 280–285.
Rasmusson XD, Zonderman AB, Kawas C, Resnick SM. Effects of age and dementia on the Trail Making Test. Clin Neuropsychol 1998; 12: 169–178.
Ferman TJ, Smith GE, Boeve BF, Graff-Radford NR, Lucas JA, Knopman DS et al. Neuropsychological differentiation of dementia with Lewy bodies from normal aging and Alzheimer's disease. Clin Neuropsychol 2006; 20: 623–636.
Giovagnoli AR, Del Pesce M, Mascheroni S, Simoncelli M, Laiacona M, Capitani E. Trail making test: normative values from 287 normal adult controls. Ital J Neurol Sci 1996; 17: 305–309.
Hamdan AC, Hamdan EML. Effects of age and education level on the Trail Making Test in a healthy Brazilian sample. Psychol Neurosci 2009; 2: 199.
Hashimoto R, Meguro K, Lee E, Kasai M, Ishii H, Yamaguchi S. Effect of age and education on the Trail Making Test and determination of normative data for Japanese elderly people: the Tajiri Project. Psychiatry Clin Neurosci 2006; 60: 422–428.
Hester RL, Kinsella GJ, Ong B, McGregor J. Demographic influences on baseline and derived scores from the trail making test in healthy older Australian adults. Clin Neuropsychol 2005; 19: 45–54.
Periáñez JA, Ríos-Lago M, Rodríguez-Sánchez JM, Adrover-Roig D, Sánchez-Cubillo I, Crespo-Facorro B et al. Trail Making Test in traumatic brain injury, schizophrenia, and normal ageing: sample comparisons and normative data. Arch Clin Neuropsychol 2007; 22: 433–447.
Seo EH, Lee DY, Kim KW, Lee JH, Jhoo JH, Youn JC et al. A normative study of the Trail Making Test in Korean elders. Int J Geriatr Psychiatry 2006; 21: 844–852.
Salthouse TA. Cognitive correlates of cross-sectional differences and longitudinal changes in trail making performance. J Clin Exp Neuropsychol 2011; 33: 242–248.
Marvel CL, Paradiso S. Cognitive and neurological impairment in mood disorders. Psychiatr Clin North Am 2004; 27: 19–viii.
Aleman A, Hijman R, Haan EHFd, Kahn RS. Memory impairment in schizophrenia: a meta-analysis. Am J Psychiatry 1999; 156: 1358–1366.
Wölwer W, Gaebel W. Impaired Trail-Making Test-B performance in patients with acute schizophrenia is related to inefficient sequencing of planning and acting. J Psychiatr Res 2002; 36: 407–416.
Heinrichs RW, Zakzanis KK. Neurocognitive deficit in schizophrenia: a quantitative review of the evidence. Neuropsychology 1998; 12: 426–445.
Gilvarry CM, Russell A, Hemsley D, Murray RM. Trail making test performance in the first degree relatives of schizophrenic and affective psychotic patients. Cogn Neuropsychiatry 2000; 5: 219–234.
Sitskoorn MM, Aleman A, Ebisch SJH, Appels MCM, Kahn RS. Cognitive deficits in relatives of patients with schizophrenia: a meta-analysis. Schizophrenia Res 2004; 71: 285–295.
Zalla T, Joyce C, Szöke A, Schürhoff F, Pillon B, Komano O et al. Executive dysfunctions as potential markers of familial vulnerability to bipolar disorder and schizophrenia. Psychiatry Res 2004; 121: 207–217.
Buyske S, Bates ME, Gharani N, Matise TC, Tischfield JA, Manowitz P. Cognitive traits link to human chromosomal regions. Behav Genet 2005; 36: 65–76.
Knowles EEM, Carless MA, de Almeida MAA, Curran JE, McKay DR, Sprooten E et al. Genome-wide significant localization for working and spatial memory: Identifying genes for psychosis using models of cognition. Am J Med Genet B Neuropsychiatr Genet 2014; 165: 84–95.
Vasilopoulos T, Franz CE, Panizzon MS, Xian H, Grant MD, Lyons MJ et al. Genetic architecture of the Delis-Kaplan Executive Function System Trail Making Test: evidence for distinct genetic influences on executive function. Neuropsychology 2012; 26: 238–250.
Ibrahim-Verbaas CA, Bressler J, Debette S, Schuur M, Smith AV, Bis JC et al. GWAS for executive function and processing speed suggests involvement of the CADM2 gene. Mol Psychiatry 2016; 21: 189–197.
Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151). Mol Psychiatry 2016; 21: 758–767.
Davies G, Armstrong N, Bis JC, Bressler J, Chouraki V, Giddaluru S et al. Genetic contributions to variation in general cognitive function: a meta-analysis of genome-wide association studies in the CHARGE consortium (N=53 949). Mol Psychiatry 2015; 20: 183–192.
Lee T, Mosing MA, Henry JD, Trollor JN, Ames D, Martin NG et al. Genetic influences on four measures of executive functions and their covariation with general cognitive ability: the Older Australian Twins Study. Behav Genet 2012; 42: 528–538.
Goul WR, Brown M. Effects of age and intelligence on Trail Making Test performance and validity. Percept Motor Skills 1970; 30: 319–326.
Waldmann BW, Dickson AL, Monahan MC, Kazelskis R. The relationship between intellectual ability and adult performance on the trail making test and the symbol digit modalities test. J Clin Psychol 1992; 48: 360–363.
Steinberg BA, Bieliauskas LA, Smith GE, Ivnik RJ. Mayo’s older Americans Normative Studies: age- and IQ-adjusted norms for the Trail-Making Test, the Stroop Test, and MAE Controlled Oral Word Association Test. Clin Neuropsychol 2005; 19: 329–377.
Corrigan JD, Hinkeldey NS. Relationships between Parts A and B of the Trail Making Test. J Clin Psychol 1987; 43: 402–409.
Sanchez-Cubillo I, Perianez J, Adrover-Roig D, Rodriguez-Sanchez J. Construct validity of the Trail Making Test: role of task-switching, working memory, inhibition/interference control, and visuomotor abilities. J Int Neuropsychol Soc 2009; 15: 438.
Oosterman JM, Vogels RLC, van Harten B, Gouw AA, Poggesi A, Scheltens P et al. Assessing mental flexibility: neuroanatomical and neuropsychological correlates of the trail making test in elderly people. Clin Neuropsychol 2010; 24: 203–219.
Rabbitt P. Between-individual variability and interpretation of associations between neurophysiological and behavioral measures in aging populations: Comment on Salthouse. 2011Psychol Bull 2011; 137: 785–789.
Cox SR, MacPherson SE, Ferguson KJ, Nissan J, Royle NA, MacLullich AMJ et al. Correlational structure of ‘frontal’ tests and intelligence tests indicates two components with asymmetrical neurostructural correlates in old age. Intelligence 2014; 46: 94–106.
Delis DC, Jacobson M, Bondi MW, Hamilton JM, Salmon DP. The myth of testing construct validity using factor analysis or correlations with normal or mixed clinical populations: lessons from memory assessment. J Int Neuropsychol Soc 2003; 9: 936–946.
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015; 12: e1001779.
Hagenaars SP, Harris SE, Davies G, Hill WD, Liewald DC, Ritchie SJ et al. Shared genetic aetiology between cognitive functions and physical and mental health in UK Biobank (N=112 151) and 24 GWAS consortia. Mol Psychiatry 2016; 21: 1624–1632.
Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Artigas MS et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med 2015; 3: 769–781.
Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–913.
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 2010; 42: 565–569.
Crocker L, Algina J. Introduction to Classical and Modern Test Theory. ERIC, 1986.
Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015; 47: 291–295.
Debette S, Ibrahim Verbaas CA, Bressler J, Schuur M, Smith A, Bis JC et al. Genome-wide studies of verbal declarative memory in nondemented older people: The Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium. Biol Psychiatry 2015; 77: 749–763.
de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol 2015; 17: e1004219.
Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 2015; 47: 1228–1235.
Euesden J, Lewis CM, O'Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics 2015; 31: 1466–1468.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B (Methodol) 1995; 57: 289–300.
Wechsler DWAIS-III: Administration and Scoring Manual: Wechsler Adult Intelligence Scale Psychological Corporation: San Antonio, TX, 1997.
Wechsler D. Wechsler Memory Scale (WMS-III). Psychological Corporation: San Antonio, TX, 1997.
Raven JC, Court JH. Manual for Raven Progressive Matrices and Vocabulary Scales. Psychological Corporation: San Antonio, TX, 1993.
Gilmour AR, Gogel B, Cullis B, Thompson R, Butler DASReml User Guide Release 3.0. VSN International Ltd: Hemel Hempstead, UK 2009.
Chung S, Zhou Z, Huddleston KA, Harrison DA, Reed R, Coleman TA et al. Crooked neck is a component of the human spliceosome and implicated in the splicing process. Biochim Biophys Acta 2002; 1576: 287–297.
Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem 1999; 68: 383–424.
Starheim KK, Arnesen T, Gromyko D, Ryningen A, Varhaug JE, Lillehaug JR. Identification of the human N(alpha)-acetyltransferase complex B (hNatB): a complex important for cell-cycle progression. Biochem J 2008; 415: 325–331.
Gennarino Vincenzo A, Singh Ravi K, White Joshua J, De Maio A, Han K, Kim J-Y et al. Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 Levels. Cell 2015; 160: 1087–1098.
Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry 2011; 16: 996–1005.
Visscher PM, Yang J, Goddard ME. A Commentary on ‘Common SNPs explain a large proportion of the heritability for human height’ by Yang et al. 2010Twin Res Hum Genet 2010; 13: 517–524.
Salthouse TA. Localizing age-related individual differences in a hierarchical structure. Intelligence 2004; 32: 541–561.
Tucker-Drob EM. Differentiation of cognitive abilities across the life span. Dev Psychol 2009; 45: 1097–1118.
Ritchie SJ, Tucker-Drob EM, Cox SR, Corley J, Dykiert D, Redmond P et al. Predictors of ageing-related decline across multiple cognitive functions. Intelligence 2016; 59: 115–126.
Acknowledgments
This research was conducted using the UK Biobank Resource under application 10279. The work was undertaken in The University of Edinburgh Centre for Cognitive Ageing and Cognitive Epidemiology, part of the cross council Lifelong Health and Wellbeing Initiative (MR/K026992/1); funding from the BBSRC and Medical Research Council (MRC) is gratefully acknowledged. The Lothian Birth Cohort is supported by Age UK (Disconnected Mind Project). SRC is supported by a grant from the MRC (MR/M013111/1). WDH is supported by a grant from Age UK (Disconnected Mind Project). AMM and IJD are supported by a Wellcome Trust Strategic Award (104036/Z/14/Z). This report represents independent research part-funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of Interest
IJD is a participant in UK Biobank. All other authors declare no conflict of interest.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if thematerial is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Hagenaars, S.P., Cox, S.R., Hill, W.D. et al. Genetic contributions to Trail Making Test performance in UK Biobank. Mol Psychiatry 23, 1575–1583 (2018). https://doi.org/10.1038/mp.2017.189
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2017.189
This article is cited by
-
Effects of home-based manual dexterity training on cognitive function among older adults: a randomized controlled trial
European Review of Aging and Physical Activity (2023)
-
Genetic modifiers in rare disorders: the case of fragile X syndrome
European Journal of Human Genetics (2021)