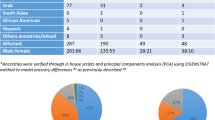
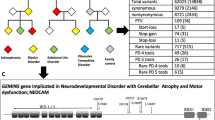
Abstract
A current focus in psychiatric genetics is detection of multiple common risk alleles through very large genome-wide association study analyses. Yet families do exist, albeit rare, that have multiple affected members who are presumed to have a similar inherited cause to their illnesses. We hypothesized that within some of these families there may be rare highly penetrant mutations that segregate with illness. In this exploratory study, the genomes of 90 individuals across nine families were sequenced. Each family included a minimum of three available relatives affected with a psychotic illness and three available unaffected relatives. Twenty-six variants were identified that are private to a family, alter protein sequence, and are transmitted to all sequenced affected individuals within the family. In one family, seven siblings with schizophrenia spectrum disorders each carry a novel private missense variant within the SHANK2 gene. This variant lies within the consensus SH3 protein-binding motif by which SHANK2 may interact with post-synaptic glutamate receptors. In another family, four affected siblings and their unaffected mother each carry a novel private missense variant in the SMARCA1 gene on the X chromosome. Both variants represent candidates that may be causal for psychotic disorders when considered in the context of their transmission pattern and known gene and disease biology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427.
Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014; 506: 185–190.
Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014; 506: 179–184.
Pocklington AJ, Rees E, Walters JT, Han J, Kavanagh DH, Chambert KD et al. Novel findings from CNVs implicate inhibitory and excitatory signaling complexes in schizophrenia. Neuron 2015; 86: 1203–1214.
Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry 2014; 204: 108–114.
McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C et al. Mosaic copy number variation in human neurons. Science 2013; 342: 632–637.
Sullivan PF, Kendler KS, Neale MC . Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 2003; 60: 1187–1192.
Bansal V, Libiger O . Fast individual ancestry inference from DNA sequence data leveraging allele frequencies for multiple populations. BMC Bioinformatics 2015; 16: 4.
International HapMap C. The International HapMap Project. Nature 2003; 426: 789–796.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498.
Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013; 11: 11 10 11–11 10 33.
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England) 2009; 25: 1754–1760.
Handsaker RE, Van Doren V, Berman JR, Genovese G, Kashin S, Boettger LM et al. Large multiallelic copy number variations in humans. Nat Genet 2015; 47: 296–303.
Layer RM, Chiang C, Quinlan AR, Hall IM . LUMPY: a probabilistic framework for structural variant discovery. Genome Biol 2014; 15: R84.
McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F . Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics (Oxford, England) 2010; 26: 2069–2070.
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29: 308–311.
Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65.
Hassel BA, Zhou A, Sotomayor C, Maran A, Silverman RH . A dominant negative mutant of 2-5 A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J 1993; 12: 3297–3304.
Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S et al. Ensembl 2014. Nucleic Acids Res 2014; 42: D749–D755.
Yang MS, Yu L, Guo TW, Zhu SM, Liu HJ, Shi YY et al. Evidence for association between single nucleotide polymorphisms in T complex protein 1 gene and schizophrenia in the Chinese Han population. J Med Genet 2004; 41: e63.
Tang W, Shi Y, Feng G, Yan L, Xing Y, Zhu S et al. Family-based association studies of the TCP1 gene and schizophrenia in the Chinese Han population. J Neural Transmission (Vienna) 2006; 113: 1537–1543.
Chu TT, Liu Y . An integrated genomic analysis of gene-function correlation on schizophrenia susceptibility genes. J Hum Genet 2010; 55: 285–292.
Athanasiu L, Mattingsdal M, Melle I, Inderhaug E, Lien T, Agartz I et al. Intron 12 in NTRK3 is associated with bipolar disorder. Psychiatry Res 2011; 185: 358–362.
Nurnberger JI Jr, Koller DL, Jung J, Edenberg HJ, Foroud T, Guella I et al. Identification of pathways for bipolar disorder: a meta-analysis. JAMA Psychiatry 2014; 71: 657–664.
Otnaess MK, Djurovic S, Rimol LM, Kulle B, Kahler AK, Jonsson EG et al. Evidence for a possible association of neurotrophin receptor (NTRK-3) gene polymorphisms with hippocampal function and schizophrenia. Neurobiol Dis 2009; 34: 518–524.
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013; 45: 580–585.
Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, Sheng M et al. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem 1999; 274: 29510–29518.
Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 1999; 23: 569–582.
Pawson T, Schlessingert J . SH2 and SH3 domains. Curr Biol 1993; 3: 434–442.
Sheng M, Kim E . The Shank family of scaffold proteins. J Cell Sci 2000; 113: 1851–1856.
Chau BN, Cheng EH, Kerr DA, Hardwick JM . Aven, a novel inhibitor of caspase activation, binds Bcl-xL and Apaf-1. Mol Cell 2000; 6: 31–40.
Stephens SH, Franks A, Berger R, Palionyte M, Fingerlin TE, Wagner B et al. Multiple genes in the 15q13-q14 chromosomal region are associated with schizophrenia. Psychiatric Genet 2012; 22: 1–14.
Thomas H, Beck K, Adamczyk M, Aeschlimann P, Langley M, Oita RC et al. Transglutaminase 6: a protein associated with central nervous system development and motor function. Amino Acids 2013; 44: 161–177.
Wang JL, Yang X, Xia K, Hu ZM, Weng L, Jin X et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain 2010; 133: 3510–3518.
Peykov S, Berkel S, Schoen M, Weiss K, Degenhardt F, Strohmaier J et al. Identification and functional characterization of rare SHANK2 variants in schizophrenia. Mol Psychiatry 2015; 20: 1489–1498.
Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet 2010; 42: 489–491.
Berkel S, Tang W, Trevino M, Vogt M, Obenhaus HA, Gass P et al. Inherited and de novo SHANK2 variants associated with autism spectrum disorder impair neuronal morphogenesis and physiology. Hum Mol Genet 2012; 21: 344–357.
Sato D, Lionel AC, Leblond CS, Prasad A, Pinto D, Walker S et al. SHANK1 deletions in males with autism spectrum disorder. Am J Hum Genet 2012; 90: 879–887.
Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet 2012; 8: e1002521.
Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 2007; 39: 25–27.
Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, Brustein E et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci USA 2010; 107: 7863–7868.
Lennertz L, Wagner M, Wolwer W, Schuhmacher A, Frommann I, Berning J et al. A promoter variant of SHANK1 affects auditory working memory in schizophrenia patients and in subjects clinically at risk for psychosis. Eur Arch Psychiatry Clin Neurosci 2012; 262: 117–124.
Alvarez-Saavedra M, De Repentigny Y, Lagali PS, Raghu Ram EV, Yan K, Hashem E et al. Snf2h-mediated chromatin organization and histone H1 dynamics govern cerebellar morphogenesis and neural maturation. Nat Commun 2014; 5: 4181.
Metzakopian E, Bouhali K, Alvarez-Saavedra M, Whitsett JA, Picketts DJ, Ang SL . Genome-wide characterisation of Foxa1 binding sites reveals several mechanisms for regulating neuronal differentiation in midbrain dopamine cells. Development 2015; 142: 1315–1324.
Yip DJ, Corcoran CP, Alvarez-Saavedra M, DeMaria A, Rennick S, Mears AJ et al. Snf2l regulates Foxg1-dependent progenitor cell expansion in the developing brain. Dev Cell 2012; 22: 871–878.
Karaca E, Harel T, Pehlivan D, Jhangiani SN, Gambin T, Coban Akdemir Z et al. Genes that affect brain structure and function identified by rare variant analyses of mendelian neurologic disease. Neuron 2015; 88: 499–513.
Koga M, Ishiguro H, Yazaki S, Horiuchi Y, Arai M, Niizato K et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum Mol Genet 2009; 18: 2483–2494.
Loe-Mie Y, Lepagnol-Bestel AM, Maussion G, Doron-Faigenboim A, Imbeaud S, Delacroix H et al. SMARCA2 and other genome-wide supported schizophrenia-associated genes: regulation by REST/NRSF, network organization and primate-specific evolution. Hum Mol Genet 2010; 19: 2841–2857.
Gershon ES, DeLisi LE, Hamovit J, Nurnberger JI Jr, Maxwell ME, Schreiber J et al. A controlled family study of chronic psychoses. Schizophrenia and schizoaffective disorder. Arch Gen Psychiatry 1988; 45: 328–336.
Proal AC, Fleming J, Galvez-Buccollini JA, Delisi LE . A controlled family study of cannabis users with and without psychosis. Schizophr Res 2014; 152: 283–288.
Jiang YH, Ehlers MD . Modeling autism by SHANK gene mutations in mice. Neuron 2013; 78: 8–27.
Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature 2012; 486: 256–260.
Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 2012; 486: 261–265.
Lopes MC, Joyce C, Ritchie GR, John SL, Cunningham F, Asimit J et al. A combined functional annotation score for non-synonymous variants. Hum Hered 2012; 73: 47–51.
Kumar P, Henikoff S, Ng PC . Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Acknowledgements
The pedigree collections were partially supported by NIMH from 1992–1999 (MHR01 44245) and more recently by Amgen 2013-present. We acknowledge the patients and families who contributed samples and made this study possible.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
LED has received funds from Amgen for clinical evaluations of multiplex families. All other co-authors are employees of Amgen. The company, however, did not influence the study design, analyses or interpretation of the results presented in this manuscript.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
Homann, O., Misura, K., Lamas, E. et al. Whole-genome sequencing in multiplex families with psychoses reveals mutations in the SHANK2 and SMARCA1 genes segregating with illness. Mol Psychiatry 21, 1690–1695 (2016). https://doi.org/10.1038/mp.2016.24
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2016.24
This article is cited by
-
Historical pursuits of the language pathway hypothesis of schizophrenia
npj Schizophrenia (2021)
-
SHANK2 mutations impair apoptosis, proliferation and neurite outgrowth during early neuronal differentiation in SH-SY5Y cells
Scientific Reports (2021)
-
Identification of SHANK2 Pathogenic Variants in a Chinese Uygur Population with Schizophrenia
Journal of Molecular Neuroscience (2021)
-
meQTL and ncRNA functional analyses of 102 GWAS-SNPs associated with depression implicate HACE1 and SHANK2 genes
Clinical Epigenetics (2020)
-
Rediscovering the value of families for psychiatric genetics research
Molecular Psychiatry (2019)