Abstract
Childhood-onset schizophrenia (COS) is a rare and severe form of the disorder, with more striking abnormalities with respect to prepsychotic developmental disorders and abnormities in the brain development compared with later-onset schizophrenia. We previously documented that COS patients, compared with their healthy siblings and with adult-onset patients (AOS), carry significantly more rare chromosomal copy number variations, spanning large genomic regions (>100 kb) (Ahn et al. 2014). Here, we interrogated the contribution of common polygenic variation to the genetic susceptibility for schizophrenia. We examined the association between a direct measure of genetic risk of schizophrenia in 130 COS probands and 103 healthy siblings. Using data from the schizophrenia and autism GWAS of the Psychiatric Genomic Consortia, we selected three risk-related sets of single nucleotide polymorphisms from which we conducted polygenic risk score comparisons for COS probands and their healthy siblings. COS probands had higher genetic risk scores of both schizophrenia and autism than their siblings (P<0.05). Given the small sample size, these findings suggest that COS patients have more salient genetic risk than do AOS.
Similar content being viewed by others
Introduction
With recent advances in genomic microarrays, identifying common genetic variants for complex traits have become a reality through genome-wide association studies (GWAS).1 For psychiatric disorders, identification of causal gene signatures has remained unclear; however, because each genetic variant or single nucleotide polymorphism (SNP) accounts for such a small proportion of the variance, necessitating very large samples.2, 3, 4 The Psychiatric Genomic Consortium (PGC) was created in order to conduct large-scale mega-analyses of GWAS data for five major psychiatric disorders, including ADHD, autism, bipolar, major depressive disorder and schizophrenia.5, 6, 7, 8, 9 Meta-analyses for these psychiatric disorders, on the one hand, confirmed many previous GWAS findings and, on the other hand, added previously unrecognized risk common variants to the list.5, 6, 7, 8, 9 However, the statistical stringency of multiple test corrections necessary for GWAS has limited studies of smaller samples.
Childhood-onset schizophrenia (COS), defined by the onset of psychotic symptoms before age 13, is a rare and severe form of the disorder. Stratifying by age of onset has been useful across all of medicine and, in particular, for identifying causal genetic variants.10, 11 We previously showed that COS patients carry a higher rate of large (>100 kb) and rare (<0.1% in controls) copy number variations that interrupt genes in pathways of neurodevelopment and regulation than do their healthy siblings or adult-onset patients (AOS).12, 13 In addition, we showed an unexpected relationship. In order to further understand the genetic susceptibility for COS, here we examined the contribution of common polygenic variation in our COS samples. In addition, we evaluated polygenic risk scores and their association with published GWAS of schizophrenia in the PGC. Finally, because COS shows a high rate of prepsychotic neurodevelopmental dysfunction,14 we also examined polygenic risk for autism in our COS sample.
Materials and Methods
Patient population
Participants were recruited as part of an ongoing National Institute of Mental Health study of COS. Patients meeting DSM-IIIR/DSM-IV criteria for schizophrenia with documented onset of psychosis before age 13 were recruited nationally. Patients and their available first-degree relatives were interviewed for lifetime and current psychiatric disorders using structured psychiatric interviews and Autism Symptom Questionnaire.15, 16 Diagnosis was confirmed with inpatient medication-free observation. A total of 600 patients were screened from which 361 were admitted for further observation. This study was approved by the Institutional Review Board of The National Institute of Mental Health. All participants provided written assent/consent with written informed consent from a parent or legal guardian for minors. A more detailed description of the inclusion protocol has been described elsewhere.13
SNP genotyping and quality control
Details of the sample collection, processing and quality controls have been described elsewhere.13 After quality control process, the remained SNP set was then used to calculate principle components through EIGENSOFT1 (www.hsph.harvard.edu/alkes-price/software) to assess for population stratification and adjust for this in the later analysis. The data were then imputed to HapMap version 3 using the MACH software (Center for Statistical Genetics, University of Michigan; http://www.sph.umich.edu/csg/abecasis/MACH)17 with the default setting and a quality control (R>0.8).
Selection of SNPs and risk score calculation
Two sets of SNPs were generated from published GWAS of schizophrenia (PGC-SCZ) and autism spectrum disorder (PGC-ASD) in the Psychiatric Genetic Consortium.5 The first set consisted of top 80 significant SNPs (P<4 × 10−5) from PGC-SCZ for the association test. Another three lists of SNPs were derived from risk markers at significance thresholds of P<0.4, P<0.2 and P<0.1. Polygenic scores were calculated using Plink V. 1.07 (http://pngu.mgh.harvard.edu/~purcell/plink)18 using the method described by Purcell et al.19 In addition, we scored our samples using the most recent 108 schizophrenia risk loci from the PGC.20 Briefly, estimates of the log of the odds ratios of association tests were obtained from the PGC-SCZ or PGC-ASD, and for each SNP, the log of odds ratio of the an allele was multiplied by (0, 1 or 2) depending on the number of reference alleles than an individual carries. The total polygenic score is a sum of across SNPs. Analyses were performed separately for score alleles derived from PGC-SCZ and PGC-ASD data sets.
Statistical analyses
Family-based association test was used to examine the association between selected SNPs and COS using PBAT software (http://www.hsph.harvard.edu/fbat/default.html).21
For the comparisons of siblings and probands’ polygenic scores, we also used linear mixed models in SAS version 9.3 (SAS Institute, Cary, NC, USA), adjusting for family membership and for the first five principal components in order to take into account possible population stratification. Nagelkerke’s pseudo R2 was used to assess the variance explained. We investigated if a polygenic score was associated with COS status. A one-tailed test was applied under a directional hypothesis that expects higher scores to be associated with an increased risk of disease.
Results
A total of 130 COS probands, 210 of their parents and 103 of their full healthy siblings passed all quality control. Demographic characteristics of COS probands are shown in Table 1. Due to the diverse ethnicity background, we performed analyses of all possible subjects, and only white Caucasians, separately.
Selected SNP analysis
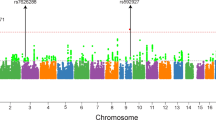
Only COS cases, their biological parents, and full siblings (443 individuals) were included in this analysis. This small sample size had under 5% of power to detect any association (effect size=2) in 1 m genome-wide scan, calculated in PBAT. Due to this lack of statistical power to conduct genome-wide association tests, we selected the top 80 significant SNPs nominated by PGC-SCZ in order to test any association with COS. This design allowed at most 50% (effect size=2) of statistical power. Table 2a showed six significant SNPs (P<0.05) before the Bonferroni correction for the multiple tests using all of possible subjects. Two markers, rs9662700, on chromosome 1 and rs17512836, on chromosome 18 survived after correction for multiple comparisons (P<6.25 × 10−4). As shown in Table 2b, from an analysis using only Caucasian (208 individuals), there were eight markers before the correction, but only one marker, rs17512836, remained significant after correction. The SNP rs17595731 is intronic for transcription factor 4 (TCF4) gene. Interestingly, TCF4 has also been reported as a risk gene for bipolar disorder22 and schizophrenia,5, 8, 23 and common TCF4 variants are involved in psychosis pathology, probably related to abnormal neurodevelopment.24
Polygenic score analysis
For this analysis, 130 COS probands and 103 their healthy siblings were included. Seventy-one probands have at least one healthy full sibling, and the mixed effect model was used in order to control the random effect due to the family membership. In the analyses based on the PGC-SCZ data, polygenic scores for schizophrenia in COS probands were higher than those in sibling group (P<0.001). The polygenic scores based on 108 risk loci in Schizophrenia Working Group of the Psychiatric Genomics20 also showed that COS patients had significantly higher score than their healthy siblings (P=0.025, R2=0.0552).
In the analyses based on the PGC-ASD, however, the scores of COS probands were also significantly higher than healthy sibling group, but at only one GWAS significance threshold (PT<0.4) (see Table 3).
We also examined the relationship between autism spectrum questionnaire score and polygenetic scores driven by Autism GWAS of PGC as 18.5% of our COS probands (n=24) had autism spectrum disorders and many of rare copy number variation abnormalities overlap as risk for autism and schizophrenia.13 We found that the polygenetic score driven by autism risk alleles had no correlation with autism spectrum questionnaire score (r=−0.06881, P=0.617). On the basis of pseudo R2, the polygenic risk score accounted for 7.5% when PT=0.4; analyses using only Caucasians showed the same patterns of results.
Discussion
In this study, we found that the polygenic risk score for schizophrenia, created by using 80 selected genetic common variants from the schizophrenia GWAS in PGC, effectively predicted COS status. Vorstman et al.6 failed to show that the polygenic score derived from adult-onset schizophrenia case–control data set could differentiate autism cases (n=2 736) from controls. This study is also the first demonstration of significance overlap in polygenic susceptibility to autism and early-onset schizophrenia, although the relationship was detectable only using the most liberal significance threshold, PT<0.4. On the other hand, this observation is consistent with our previous reports that autism and COS share several rare copy number variations as risk factors.13 These findings suggest that COS may share additional common risk markers with autism.
We replicated the association between rs17512836 (intron 3 in TCF4), located on chromosome 18q21, and COS risk. TCF4 is highly expressed in the brain, and has a role in neurodevelopment, interacting with class II bHLH transcription factors Math1, HASH1 and neuroD2. The Ca(2+) sensor protein calmodulin interacts with the DNA binding domain of TCF4, inhibiting transcriptional activation.25, 26 Because of its interaction with calmodulin and involvement in calcium signaling, TCF4 may also have direct functional neuronic effects.26 Although family-based association can be controlled for population stratification, COS samples were ethnically heterogeneous. Moreover, our sample, is necessarily small, given the rarity of this subgroup of patients. A definitive statement regarding risk loci and effect size cannot be made conclusively until replication studies are performed.
The estimated variances using R2 based on the schizophrenia risk variants in this sample (range 5.5 ~18.5%) are much stronger than that for the polygenic score representing later-onset schizophrenia (~6%)5 or bipolar disorder (~3%).27 These findings suggest that COS patients have a more salient genetic risk with respect to common variants than do adult-onset patients.
References
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D . Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
Moonesinghe R, Khoury MJ, Liu T, Ioannidis JP . Required sample size and nonreplicability thresholds for heterogeneous genetic associations. Proc Natl Acad Sci USA 2008; 105: 617–622.
Visscher PM, Brown MA, McCarthy MI, Yang J . Five years of GWAS discovery. Am J Hum Genet 2012; 90: 7–24.
Chapman K, Ferreira T, Morris A, Asimit J, Zeggini E . Defining the power limits of genome-wide association scan meta-analyses. Genet Epidemiol 2011; 35: 781–789.
Schizophrenia Psychiatric Genome-Wide Association Study Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet 2011; 43: 969–976.
Vorstman JA, Anney RJ, Derks EM, Gallagher L, Gill M, de Jonge MV et al. No evidence that common genetic risk variation is shared between schizophrenia and autism. Am J Med Genet B Neuropsychiatr Genet 2013; 162B: 55–60.
Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet 2013; 45: 984–994.
Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D et al. Common variants conferring risk of schizophrenia. Nature 2009; 460: 744–747.
Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch KP et al. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry 2010; 49: 884–897.
Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 1990; 250: 1684–1689.
Murrell J, Farlow M, Ghetti B, Benson MD . A mutation in the amyloid precursor protein associated with hereditary Alzheimer's disease. Science 1991; 254: 97–99.
Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 2008; 320: 539–543.
Ahn K, Gotay N, Andersen TM, Anvari AA, Gochman P, Lee Y et al. High rate of disease-related copy number variations in childhood onset schizophrenia. Mol Psychiatry 2014; 19: 568–572.
Addington AM, Rapoport JL . The genetics of childhood-onset schizophrenia: when madness strikes the prepubescent. Curr Psychiatry Rep 2009; 11: 156–161.
Kumra S, Frazier JA, Jacobsen LK, McKenna K, Gordon CT, Lenane MC et al. Childhood-onset schizophrenia. A double-blind clozapine-haloperidol comparison. Arch Gen Psychiatry 1996; 53: 1090–1097.
Shaw P, Sporn A, Gogtay N, Overman GP, Greenstein D, Gochman P et al. Childhood-onset schizophrenia: a double-blind, randomized clozapine-olanzapine comparison. Arch Gen Psychiatry 2006; 63: 721–730.
Li Y, Willer C, Sanna S, Abecasis G . Genotype imputation. Annu Rev Genomics Hum Genet 2009; 10: 387–406.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC et alInternational Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752.
Schizophrenia Working Group of the Psychiatric Genomics Consotrium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427.
Laird NM, Horvath S, Xu X . Implementing a unified approach to family-based tests of association. Genet Epidemiol 2000; 19: S36–S42.
Del-Favero J, Gestel SV, Borglum AD, Muir W, Ewald H, Mors O et al. European combined analysis of the CTG18.1 and the ERDA1 CAG/CTG repeats in bipolar disorder. Eur J Hum Genet 2002; 10: 276–280.
Steinberg S, de Jong S, Irish Schizophrenia Genomics Consortium, Andreassen OA, Werge T, Borglum AD et al. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet 2011; 20: 4076–4081.
Wirgenes KV, Sonderby IE, Haukvik UK, Mattingsdal M, Tesli M, Athanasiu L et al. TCF4 sequence variants and mRNA levels are associated with neurodevelopmental characteristics in psychotic disorders. Trans Psychiatry 2012; 2: e112.
Mudge J, Miller NA, Khrebtukova I, Lindquist IE, May GD, Huntley JJ et al. Genomic convergence analysis of schizophrenia: mRNA sequencing reveals altered synaptic vesicular transport in post-mortem cerebellum. PloS One 2008; 3: e3625.
Navarrete K, Pedroso I, De Jong S, Stefansson H, Steinberg S, Stefansson K et al. TCF4 (e2-2; ITF2): a schizophrenia-associated gene with pleiotropic effects on human disease. Am J Med Genet B Neuropsychiatr Genet 2013; 162B: 1–16.
Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43: 977–983.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Ahn, K., An, S., Shugart, Y. et al. Common polygenic variation and risk for childhood-onset schizophrenia. Mol Psychiatry 21, 94–96 (2016). https://doi.org/10.1038/mp.2014.158
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2014.158
This article is cited by
-
Progress and Implications from Genetic Studies of Bipolar Disorder
Neuroscience Bulletin (2024)
-
Age-related change in cortical thickness in adolescents at clinical high risk for psychosis: a longitudinal study
European Child & Adolescent Psychiatry (2023)
-
Impact on the Risk and Severity of Childhood Onset Schizophrenia of Schizophrenia Risk Genetic Variants at the DRD2 and ZNF804A Loci
Child Psychiatry & Human Development (2023)
-
The interaction between first-episode drug-naïve schizophrenia and age based on gray matter volume and its molecular analysis: a multimodal magnetic resonance imaging study
Psychopharmacology (2023)
-
Neurexins in autism and schizophrenia—a review of patient mutations, mouse models and potential future directions
Molecular Psychiatry (2021)