Abstract
Sinonasal undifferentiated carcinoma is a rare and aggressive malignancy. Sinonasal undifferentiated carcinoma has long been considered a diagnosis of exclusion; to date, the molecular pathogenetic basis for sinonasal undifferentiated carcinoma is unknown. To identify potential oncogenic drivers in sinonasal undifferentiated carcinoma, targeted next-generation sequencing of 300 cancer-related genes was performed on 11 cases of sinonasal undifferentiated carcinoma. We identified IDH2 R172X mutations in 55% of sinonasal undifferentiated carcinomas including R172S, R172T, and R172M. Multispecific mutant IDH1/2 immunohistochemistry was performed and identified mutant-specific protein expression in all cases with available tissue: 3/3 sinonasal undifferentiated carcinomas with R172 mutations were positive and 4/4 wild-type cases were negative. Review of sequencing data for our institutional head and neck cohorts (n=412) confirmed the absence of IDH-activating mutations in other tumor types. Alterations in the IDH2-wild-type sinonasal undifferentiated carcinomas included SMARCA4 loss-of-function with confirmed loss of immunohistochemical expression, NOTCH1 gain-of-function, and TET2 loss-of-function. We demonstrate that the majority of histologically defined sinonasal undifferentiated carcinomas are characterized by IDH2 R172X mutations and overexpression of mutant protein. IDH2 R172X mutations are specific to sinonasal undifferentiated carcinoma among carcinomas of the head and neck, confirming this tumor type as a distinct clinicopathologic entity. These findings have significant implications for diagnosis and therapy with IDH inhibitors for patients with this rare and poorly understood tumor.
Similar content being viewed by others
Main
Sinonasal undifferentiated carcinoma is a poorly characterized, rare, and aggressive epithelial malignancy of the sinonasal tract. Sinonasal undifferentiated carcinoma most commonly afflicts older adults, although a wide age range has been observed.1 Sinonasal undifferentiated carcinoma presents as a fast growing, locally destructive and invasive tumor, often extending into the cranial vault.2 Mortality is high, with an overall 2-year survival rate <50% despite the multimodal therapy.3, 4 As sinonasal undifferentiated carcinoma was first described in 1986,5 few insights have been made into its pathogenesis, and no current standardized treatment regimen exists. Tumors originally considered to represent sinonasal undifferentiated carcinoma are likely heterogeneous, as genomically defined carcinomas have been subsequently identified within this ‘undifferentiated’ group, including NUT-midline carcinoma,6, 7 nasopharyngeal carcinoma,8, 9 HPV-related carcinomas,10 and SMARCB1 (INI1)-deficient carcinoma.11, 12
Uniform pathologic diagnostic criteria for sinonasal undifferentiated carcinoma are not well established; this tumor type has long been considered a diagnosis of exclusion once epithelial differentiation is confirmed and other entities have been excluded. Tumors express keratins and EMA but no other differentiation markers with rare exceptions, including focal staining for synaptophysin, chromogranin, S-100, and p63/p40.5, 9, 13, 14, 15 Thus far, no recurrent genetic alterations have been identified in sinonasal undifferentiated carcinoma16 and no specific diagnostic markers exist.
Our aim was to evaluate a cohort of sinonasal undifferentiated carcinoma using large panel-targeted next-generation sequencing to identify recurrent genetic alterations, in an effort to better define this poorly understood and lethal tumor.
Materials and methods
Tumor Samples
Eleven cases of sinonasal undifferentiated carcinoma diagnosed between 2007 and 2016 were retrospectively identified in the surgical pathology archives. Hematoxylin and eosin- and immunohistochemically stained slides were reviewed by two specialists in head and neck pathology (VYJ and JFK) for diagnostic confirmation, excluding known mimics when possible. For example, NUT-midline carcinomas, SMARCB1 (INI1)-deficient carcinomas, and nasopharyngeal undifferentiated carcinomas (positive by in situ hybridization for Epstein–Barr virus-encoded RNA; EBER) were excluded, as were tumors that showed strong and diffuse p63 or p40 immunoreactivity. Morphologic features were reviewed blinded to immunohistochemical and sequencing results. This study was performed with approval by the Institutional Review Board at Brigham and Women's Hospital and Dana Farber Cancer Institute (Boston, MA, USA).
Sequencing Analysis
Next-generation sequencing was performed at Brigham and Women's Hospital. The OncoPanel assay surveys DNA sequences of all coding exons of 300 cancer-related genes and 113 introns across 35 genes for rearrangement detection using massively parallel sequencing (see Supplementary Table 1 for a complete list of targeted genes). DNA was extracted from formalin-fixed paraffin-embedded tumor samples using QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA), and then sheared into fragments of 250 bp average size with a sonicator (Covaris, Woburn, MA, USA). TruSeq Sample Preparation Kit (Illumina, San Diego, CA, USA) was used to construct DNA libraries. Pools of single-stranded RNA probes were designed and synthesized as part of the Agilent SureSelect Hybrid Capture Kit (Agilent, Santa Clara, CA, USA) and used to bait DNA of interest from the sample library. Selected DNA was quantified, normalized, and pooled, and then sequenced with an Illumina HiSeq 2500 sequencer. Sequence data were demultiplexed and aligned using the Burrows-Wheeler Aligner software tool. The MuTect and GATK software was used to detect single-nucleotide variants and small insertion-deletions (indels),17, 18 and copy number variations were detected using RobustCNV, a tool that is based on the calculation of log2 ratio of read counts of individual specimens against a panel of normal tissues. BreakMer analysis was performed to detect large structural variations as described previously.19 Any calls present at >0.1% in the Exome Variant Server (http://evs.gs.washington.edu/EVS/) were automatically filtered, with a rescue for any variants also present in Cosmic (http://cancer.sanger.ac.uk/cosmic) two or more times. All single-nucleotide variants, copy number variations, and translocation calls were manually reviewed and annotated for pathogenetic significance (LMS). Alterations were classified on a 4-tier scheme according to clinical relevance: Tier 1, actionable target using a Food and Drug Administration-approved therapy; Tier 2, prognostic implications or clinical trial drug target; Tier 3, inferred biological relevance; and Tier 4, variant of unknown significance.
Sequencing data from an institutional cohort of head and neck tumors (n=412; including three sinonasal undifferentiated carcinomas from this cohort) assayed by OncoPanel under a consented cancer sequencing program (PROFILE, DFCI IRB nos 11–104)20 were reviewed using an institutional instance of cBioPortal.21
Immunohistochemistry and In Situ Hybridization
IDH2 immunohistochemistry was performed on formalin-fixed paraffin-embedded whole sections from seven cases with available tumor tissue following pressure cooker antigen retrieval (pH 6.1; Target Retrieval Solution; Dako, Carpinteria, CA, USA) using a mouse monoclonal antibody for IDH1/2 mutant R132/172 (clone MsMab-1; 1:100 dilution; Millipore, Darmstadt, Germany) with Novolink Detection System (Leica, Buffalo Grove, IL, USA). A colorectal adenocarcinoma with known IDH2 R172S mutation was used as a positive control. Immunohistochemistry results were scored blinded to the IDH2 mutational status. Positive staining was characterized by granular cytoplasmic staining in tumor cells. The other antibodies and conditions for the immunohistochemical studies performed for this cohort are detailed in Table 1. The Envision Plus detection System (Dako, Carpinteria, CA, USA) was used for all other antibodies besides IDH1/2.
In situ hybridization for EBER was performed on a Ventana Benchmark XT autostainer (Ventana, Tucson, AZ, USA) using an 1 DNP probe (Ventana; cat. no. 760-1209A) and ISH iView Blue Plus anti-DNP Detection System (Ventana).
Statistics
Survival time calculation was performed using Stata/IC 12.1 (StataCorp LP, College Station, TX, USA).
Results
Clinical and Pathologic Features
The clinical characteristics of the cohort are summarized in Table 2. Three cases were diagnosed in pathologic consultation only and had limited clinical data available. The cohort consisted of six men and five women, aged 41–75 years (median 64), and included six neversmokers. Sampled tumor sites were nasal (n=5), nasopharynx (n=1), maxillary sinus (n=1), ethmoid sinus (n=2), sphenoid sinus (n=1), and pleural effusion with metastasis from a maxillary sinus primary (n=1).
All sinonasal undifferentiated carcinomas showed sheet-like and nested growth of medium-to-large cells with hyperchromatic and vesicular nuclei, prominent nucleoli, and scant-to-moderate palely eosinophilic cytoplasm (Figure 1). All cases had necrosis and brisk mitotic activity (median count of 18.5 per 10 high-power fields (range, 5–47)). Lymphovascular and perineural invasion were frequent. No evidence of squamous differentiation (keratinization) or associated surface squamous dysplasia was observed; duct or gland formation and rhabdoid cytoplasmic inclusions were absent in all cases. All tumors showed strong and diffuse keratin expression (11/11). Rare reactivity was seen for p63/p40 (1/10), synaptophysin (2/10), chromogranin (3/9), and S-100 (2/9). Negative stains included desmin (0/7), NUT (0/8), and EBER (0/6); expression of SMARCB1 (INI1) was retained (7/7), and SMARCA4 (BRG1) expression was lost in 1/7 (see below). Table 3 summarizes the pathologic features and immunohistochemical results for the cohort.
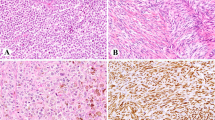
Sinonasal undifferentiated carcinomas with IDH2 R172X mutations. Case 6: (a) Right nasal mass biopsy, hematoxylin and eosin (H&E) stain; (b) IDH2 c.516G>C (p.R172S) (Integrated Genome Viewer, Broad Institute, Cambridge, MA, USA); (c) Immunohistochemistry for IDH1/2 R132/172 shows strong and diffuse granular cytoplasmic staining. Case 3: (d) Right ethmoid mass biopsy, H&E; (e) IDH2 c.515G>T (p.R172M) (Integrated Genome Viewer); (f) immunohistochemistry for IDH1/2 R132/172 shows weak-to-moderate multifocal granular cytoplasmic staining. Case 5: (g) nasal mass biopsy, H&E; (h) IDH2 c.516G>C (p.R172S) (Integrated Genome Viewer); (i) immunohistochemistry for IDH1/2 R132/172 shows moderate to strong and diffuse granular cytoplasmic staining. All photomicrographs at x400 magnification.
Next-Generation Sequencing Results
The median overall sequencing coverage per case was 384 reads (range, 126–610), and the median percentage of sequences achieving >30-fold coverage was 99% (range, 97–99%). The median number of single-nucleotide variants per tumor using this targeted panel was 3.1 per megabase (range 1.5–4.1 per Mb). Figure 2 summarizes the key genomic findings in the 11 cases of sinonasal undifferentiated carcinoma. A complete list of identified single-nucleotide variants and structural variants is available in Supplementary Table 2.
IDH2 mutations at the known hotspot R172 were identified in 6 of 11 (55%) sinonasal undifferentiated carcinomas, including R172S (n=4), R172T (n=1), and R172M (n=1) (Figure 1). Review of sequencing data for 409 other head and neck malignancies confirmed the absence of IDH-activating mutations in all other tumor types (see Supplementary Table 3), including 6 NUT-midline carcinomas, 4 nasopharyngeal carcinomas, 5 neuroendocrine carcinomas, 3 olfactory neuroblastomas, and 319 squamous cell carcinomas (including 11 primary sinonasal tumors).
Five sinonasal undifferentiated carcinomas were IDH2-wild type. One case had a NOTCH1 PEST domain insertion mutation leading to premature truncation (Case 7); this is a documented mechanism of NOTCH pathway activation in other tumor types.22 A SMARCA4 splice site variant and two concomitant APC loss-of-function variants (Figure 2 and Supplementary Table 2) were detected in Case 8; SMARCA4 loss of expression was confirmed by immunohistochemistry (Figure 3). A BAP1 splice site mutation along with single-copy deletion of BAP1 was observed in Case 9. Case 11 had a TET2 exon 3 frameshift mutation.
Diverse concomitant oncogenic alterations, including in PIK3CA, MTOR, SOX2, and SOX9 were also identified in the IDH2-mutated tumors. Loss-of-function alterations likely contributing to cell cycle dysregulation, including in TP53, RB1, and CDKN2A, were identified in three IDH2-mutated cases and one IDH2-wild-type case (Figure 2 and Supplementary Table 2). Loss-of-function alterations of multiple genes involved in epigenetic regulation (including ARID1A, ARID2, CREBBP, KMT2A, KMT2D, SETD2, TET2) were identified in 4/6 IDH2-mutated tumors and 8/11 overall (Figure 2 and Supplementary Table 2). In contrast to acute myeloid leukemia,23 TET2 mutations were identified both together with, and exclusive of, IDH2 mutations (Figure 2).
Copy number variant profiles were available in nine cases; copy number variant data could not be interpreted for Cases 10 and 11 because of low tumor content. Recurrent low copy gains were identified at 1q21–1q44 (5/6 IDH2-mutated sinonasal undifferentiated carcinomas; 6/9 overall), 8q13.1–8q24.3 (4/6 IDH2-mutated sinonasal undifferentiated carcinomas; 6/9 overall), and 17q23.2–17q24.3 (4/6 IDH2-mutated sinonasal undifferentiated carcinomas; 5/9 overall), including SOX9 amplification in two IDH2-mutated cases. Copy number variations at the chromosomal arm level are summarized in Figure 4.
Copy number alterations as determined by targeted next-generation sequencing in 11 cases of sinonasal undifferentiated carcinoma. Dotted lines indicate centromere position, and solid lines demarcate chromosomes. Light red=low copy gain; dark red=amplification (estimated ≥6 copies); light blue=single-copy deletion; dark blue=homozygous deletion. Tumor content in Cases 10 and 11 was too low for copy number change detection. Cases 1–6 represent IDH2-mutated tumors.
IDH1/2 Immunohistochemistry
IDH1/2 immunohistochemistry was performed in seven samples with available remaining tissue. Three of three sinonasal undifferentiated carcinomas with R172 mutations showed cytoplasmic staining for IDH1/2. Two of these tumors had R172S and showed moderate-to-strong multifocal-to-diffuse reactivity (Figure 1). One tumor harbored R172M and showed weak-to-moderate multifocal staining (Figure 1). No staining was observed in 4/4 IDH2-wild-type sinonasal undifferentiated carcinomas (Figure 5).
Negative IDH1/2 R132/172 immunohistochemistry in IDH2-wild-type sinonasal undifferentiated carcinoma. Case 7: (a) Sinonasal undifferentiated carcinoma, right maxillary sinus, hematoxylin and eosin stain; (b) negative staining by IDH1/2 R132/172 immunohistochemistry. Case 9: (c) Sinonasal undifferentiated carcinoma, left ethmoid sinus, hematoxylin and eosin stain; (d) negative staining by IDH1/2 R132/172 immunohistochemistry. Case 10: (e) Sinonasal undifferentiated carcinoma, nasopharynx, hematoxylin and eosin stain; (f) negative staining by IDH1/2 R132/172 immunohistochemistry. Case 11: (g) Metastatic sinonasal undifferentiated in a cell block from a pleural effusion (from a right maxillary sinus primary), hematoxylin and eosin stain. (h) Negative staining by IDH1/2 R132/172 immunohistochemistry. All photomicrographs at x400 magnification.
Clinical and Pathologic Features of Patients with IDH2-mutated and -Wild-Type Sinonasal Undifferentiated Carcinomas
Comparisons of the clinical features of the patients with IDH2-mutated and IDH2-wild-type tumors are limited by the small cohort size, thus no statistically robust conclusions can be drawn; however, the patients with IDH2-mutated tumors were younger (median 57 years) versus IDH2-wild-type (median 71 years). All patients presented with similar stages of disease; eight patients with known clinical data underwent radiation and chemotherapy, and surgical resection was performed for 1/4 patients with IDH2-mutated tumors and 3/4 IDH2-wild-type patients. Overall median survival for the cohort is 23 months; small numbers and limited follow-up time preclude comparison of outcomes in IDH2-mutated and -wild-type cohorts.
No morphologic or immunophenotypic differences were detected between IDH2-mutated and wild-type tumors.
Discussion
Sinonasal undifferentiated carcinoma is a rare aggressive neoplasm appearing as an undifferentiated or poorly differentiated epithelial malignancy. We performed an analysis of sinonasal undifferentiated carcinoma using targeted next-generation sequencing of 300 oncogenes and tumor suppressors. Our study demonstrates that sinonasal undifferentiated carcinomas, when defined based on morphology and the presence of strong keratin expression per traditional criteria, are genomically heterogeneous but enriched for activating IDH2 mutations. Notably, all IDH2 mutations occur at the known hotspot R172, the identification of which creates a novel, genomically defined subset of sinonasal carcinomas. Examination of the genomic sequences of over 400 other tumor types including morphologic mimics indicates that IDH2 mutations are specific to sinonasal undifferentiated carcinoma in the differential diagnosis with other head and neck carcinomas.
The identification of IDH2 mutations has significant potential implications for management of sinonasal undifferentiated carcinoma. Isocitrate dehydrogenase catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG). Mutations in IDH1 at R132 and IDH2 at R172 alter the activity of the IDH1 and IDH2 enzymes, catalyzing the reduction of α-KG to 2-hydroxyglutarate (2HG). 2HG is an oncometabolite that is structurally analogous to α-KG and can interfere with histone demethylation mediated by a family of α-KG-dependent enzymes.24 The oncogenic role of mutant IDH1 and IDH2 has been demonstrated in several human malignancies marked by a hypermethylated phenotype and blocked cellular differentiation, including subsets of acute myeloid leukemia and glioma, as well as chondrosarcomas.25, 26, 27 Based on evidence that mutant IDH1/2 inhibitors can induce differentiation in acute myeloid leukemia and gliomas,28, 29 IDH1/2 mutations are currently being investigated as biomarkers of response to IDH inhibitors. The identification of loss-of-function mutations in the α-KG-dependent DNA demethylase TET2, including in an IDH2-wild-type tumor, suggests that epigenetic dysregulation (eg, hypermethylation or impaired demethylation) is a common feature in sinonasal undifferentiated carcinoma.23 Further studies are needed to confirm this hypothesis.
Although some recurrent copy number alterations were observed, including low gains on chromosomes 1, 8, and 17, no pattern of copy gains and losses clearly distinguished IDH2-mutant and wild-type tumors. There was variability in the number of copy number variants even within the IDH2-mutated group; some tumors had relatively few gains and losses (Case 4), whereas multiple high-level amplifications (including SOX2, ETV6, SOX9) and a homozygous deletion (NF2) were observed in Case 6. The presence of copy number heterogeneity across the IDH2-mutated sinonasal undifferentiated carcinomas suggests the presence of differing levels of genomic instability, which may have implications for differential prognosis and response to therapy even within this genetically defined set of tumors.
The identification of IDH2 R172X mutations also has important diagnostic implications for this tumor type, formerly considered a diagnosis of exclusion. Indeed, a subset of cases can be identified by immunohistochemistry using a multispecific antibody for mutant IDH1/2 R132/172. However, this antibody has been shown to lack sensitivity for R172T;30 therefore, at this time-sequencing methodologies may be necessary to detect the range of IDH2 variants as well as other potentially targetable alterations in histologically similar tumors.
The differential diagnosis of sinonasal undifferentiated carcinoma can be challenging and requires ancillary testing to exclude morphologic mimics. Previous series and diagnoses of sinonasal undifferentiated carcinoma likely encompass a heterogeneous group of tumors that has been narrowed down over the past several decades with the increasing recognition of pathogenetically defined tumor types and the development of diagnostic biomarkers. Immunohistochemistry, in situ hybridization, and molecular testing can now identify many specific entities within keratin-positive poorly differentiated or undifferentiated malignancies in the sinonasal tract. The differential diagnosis is broad, and includes squamous cell carcinoma and its variants (including HPV-associated carcinoma, nasopharyngeal carcinoma, and NUT-midline carcinoma) and SMARCB1 (INI1)-deficient sinonasal carcinoma, as well as neuroendocrine carcinoma, olfactory neuroblastoma, and salivary neoplasms such as the solid variant of adenoid cystic carcinoma. Nasopharyngeal carcinoma is typically associated with a dense lymphocytic infiltrate, and appears as an undifferentiated carcinoma with sparse (if any) histologic features of squamous differentiation; p63/p40 can confirm squamous differentiation and the diagnosis can be confirmed by in situ hybridization to detect Epstein–Barr virus-encoded mRNA.8, 9 NUT-midline carcinoma, defined by rearrangements of the NUT gene (encoded on 15q14), frequently arises in the sinonasal tract; while appearing primarily as a poorly differentiated or undifferentiated carcinoma, foci of abrupt keratinization may be present and tumor cells are positive for p63/p40. CD34 is positive in less than half of all cases. NUT immunostain will show a punctate nuclear staining pattern;7, 31 FISH can also confirm NUT gene rearrangement. SMARCB1 (INI1)-deficient sinonasal carcinoma typically has an infiltrative nested or trabecular growth pattern within a fibrous stroma, with tumor cells having round nuclei, prominent nucleoli, and varying basaloid or epithelioid appearances, and rhabdoid morphology at least focally present.11, 12, 32 SMARCB1 (INI1)-deficient sinonasal carcinoma shows variable p63/p40 staining (up to 44%) and occasional p16 reactivity, and demonstrates consistent loss of expression for SMARCB1 (INI1) secondary to SMARCB1 deletion.11, 12
Although our cohort of sinonasal undifferentiated carcinomas was enriched for IDH2 mutants, it is notable that other significant potential driver events were identified in several of the IDH2-wild-type tumors. Inactivating mutations located within the NOTCH1 PEST regulatory domain lead to increased intracellular Notch expression and may predict therapeutic benefit with γ-secretase inhibitors.33 Interestingly, similar NOTCH1 mutations have been reported in the solid variant of adenoid cystic carcinoma,34 a potential morphologic mimic of sinonasal undifferentiated carcinoma. A MYB-NFIB rearrangement was not detected in the NOTCH1-mutated tumor reported here. SMARCA4 deficiency as a driver event in tumorigenesis has been reported in a variety of undifferentiated carcinomas and sarcomas arising at other sites;35, 36, 37 to our knowledge, this phenomenon has not yet been described in head and neck malignancies, but is not unexpected given the recent recognition of SMARCB1 (INI1)-deficient sinonasal carcinoma,11, 12, 32 with which the SMARCA4-deficient sinonasal carcinoma might be classified.
While the specific association with IDH2 mutations argues that sinonasal undifferentiated carcinoma deserves recognition as an independent diagnostic entity, it remains to be determined whether mutant IDH2 sinonasal undifferentiated carcinoma represents a distinct clinicopathologic entity. The detection of other driver mutations (NOTCH1 gain-of-function, SMARCA4 loss-of-function, and TET2 loss-of-function) suggests that there may be other potential genomically defined subsets within histologically defined sinonasal undifferentiated carcinomas. Limitations of our study include a small cohort and short follow-up time. Given the potential role for targeted inhibitors, pathologists and clinicians should consider sequencing to determine IDH2 status for patients with a diagnosis of sinonasal undifferentiated carcinoma. Multi-institutional studies, including prospective identification of IDH2 mutations and use of basket trials to test the activity of mutant IDH inhibitors in patients with this very rare disease, will be required to determine the true clinical implications of this finding in sinonasal undifferentiated carcinoma. In conclusion, the presence of recurrent IDH2 R172X mutations in sinonasal undifferentiated carcinoma begins to redefine this ‘diagnosis of exclusion’ and opens up genomically driven clinical trial opportunities for patients with this rare and aggressive form of carcinoma.
References
Frierson HF. Sinonasal undifferentiated carcinoma. In: Barnes L, Eveson JW, Reichart P et al (eds). WHO Classification of Tumours: Pathology and Genetics of Head and Neck Tumours. IARC Press: Lyon, France, 2005;p 19.
Chambers KJ, Lehmann AE, Remenschneider A et al, Incidence and survival patterns of sinonasal undifferentiated carcinoma in the United States. J Neurol Surg B 2015; 76: 94–100.
Musy PY, Reibel JF, Levine PA . Sinonasal undifferentiated carcinoma: the search for a better outcome. Laryngoscope 2002; 112: 1450–1455.
Enepekides DJ . Sinonasal undifferentiated carcinoma: an update. Curr Opin Otolaryngol Head Neck Surg 2005; 13: 222–225.
Frierson HF Jr, Mills SE, Fechner RE et al, Sinonasal undifferentiated carcinoma. An aggressive neoplasm derived from schneiderian epithelium and distinct from olfactory neuroblastoma. Am J Surg Pathol 1986; 10: 771–779.
French CA, Kutok JL, Faquin WC et al, Midline carcinoma of children and young adults with NUT rearrangement. J Clin Oncol 2004; 22: 4135–4139.
Stelow EB, Bellizzi AM, Taneja K et al, NUT rearrangement in undifferentiated carcinomas of the upper aerodigestive tract. Am J Surg Pathol 2008; 32: 828–834.
Leung SY, Yuen ST, Chung LP et al, Epstein-Barr virus is present in a wide histological spectrum of sinonasal carcinomas. Am J Surg Pathol 1995; 19: 994–1001.
Jeng YM, Sung MT, Fang CL et al, Sinonasal undifferentiated carcinoma and nasopharyngeal-type undifferentiated carcinoma: two clinically, biologically, and histopathologically distinct entities. Am J Surg Pathol 2002; 26: 371–376.
Bishop JA, Guo TW, Smith DF et al, Human papillomavirus-related carcinomas of the sinonasal tract. Am J Surg Pathol 2013; 37: 185–192.
Agaimy A, Koch M, Lell M et al, SMARCB1(INI1)-deficient sinonasal basaloid carcinoma: a novel member of the expanding family of SMARCB1-deficient neoplasms. Am J Surg Pathol 2014; 38: 1274–1281.
Bishop JA, Antonescu CR, Westra WH . SMARCB1 (INI-1)-deficient carcinomas of the sinonasal tract. Am J Surg Pathol 2014; 38: 1282–1289.
Tilson MP, Bishop JA . Utility of p40 in the differential diagnosis of small round blue cell tumors of the sinonasal tract. Head Neck Pathol 2014; 8: 141–145.
Singh L, Ranjan R, Arava S et al, Role of p40 and cytokeratin 5/6 in the differential diagnosis of sinonasal undifferentiated carcinoma. Ann Diagn Pathol 2014; 18: 261–265.
Cerilli LA, Holst VA, Brandwein MS et al, Sinonasal undifferentiated carcinoma: immunohistochemical profile and lack of EBV association. Am J Surg Pathol 2001; 25: 156–163.
Gelbard A, Hale KS, Takahashi Y et al, Molecular profiling of sinonasal undifferentiated carcinoma. Head Neck 2014; 36: 15–21.
McKenna A, Hanna M, Banks E et al, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
Cibulskis K, Lawrence MS, Carter SL et al, Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013; 31: 213–219.
Abo RP, Ducar M, Garcia EP et al, BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res 2015; 43: e19.
Sholl LM, Do K, Shivdasani P et al, Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016; 1: e87062.
Cerami E, Gao J, Dogrusoz U et al, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404.
Weng AP, Ferrando AA, Lee W et al, Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004; 306: 269–271.
Figueroa ME, Abdel-Wahab O, Lu C et al, Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 2010; 18: 553–567.
Xu W, Yang H, Liu Y et al, Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011; 19: 17–30.
Dang L, White DW, Gross S et al, Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009; 462: 739–744.
Ward PS, Patel J, Wise DR et al, The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 2010; 17: 225–234.
Amary MF, Bacsi K, Maggiani F et al, IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol 2011; 224: 334–343.
Wang F, Travins J, DeLaBarre B et al, Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science 2013; 340: 622–626.
Rohle D, Popovici-Muller J, Palaskas N et al, An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 2013; 340: 626–630.
Liu X, Kato Y, Kaneko MK et al, Isocitrate dehydrogenase 2 mutation is a frequent event in osteosarcoma detected by a multi-specific monoclonal antibody MsMab-1. Cancer Med 2013; 2: 803–814.
Haack H, Johnson LA, Fry CJ et al, Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am J Surg Pathol 2009; 33: 984–991.
Wasserman JK, Dickson BC, Perez-Ordonez B et al, INI1 (SMARCB1)-deficient sinonasal carcinoma: a clinicopathologic report of 2 cases. Head Neck Pathol 2017; e-pub ahead of print.
Wang K, Zhang Q, Li D et al, PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple-negative breast cancer sensitive to a gamma-secretase inhibitor. Clin Cancer Res 2015; 21: 1487–1496.
Stoeck A, Lejnine S, Truong A et al, Discovery of biomarkers predictive of GSI response in triple-negative breast cancer and adenoid cystic carcinoma. Cancer Discov 2014; 4: 1154–1167.
Agaimy A, Daum O, Markl B et al, SWI/SNF complex-deficient undifferentiated/rhabdoid carcinomas of the gastrointestinal tract: a series of 13 cases highlighting mutually exclusive loss of SMARCA4 and SMARCA2 and frequent co-inactivation of SMARCB1 and SMARCA2. Am J Surg Pathol 2016; 40: 544–553.
Karnezis AN, Wang Y, Ramos P et al, Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J Pathol 2016; 238: 389–400.
Le Loarer F, Watson S, Pierron G et al, SMARCA4 inactivation defines a group of undifferentiated thoracic malignancies transcriptionally related to BAF-deficient sarcomas. Nat Genet 2015; 47: 1200–1205.
Acknowledgements
This work was funded by the Department of Pathology, Brigham and Women's Hospital.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Jo, V., Chau, N., Hornick, J. et al. Recurrent IDH2 R172X mutations in sinonasal undifferentiated carcinoma. Mod Pathol 30, 650–659 (2017). https://doi.org/10.1038/modpathol.2016.239
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.239
This article is cited by
-
Top IHC/ISH Hacks for and Molecular Surrogates of Poorly Differentiated Sinonasal Small Round Cell Tumors
Head and Neck Pathology (2024)
-
Molecularly defined sinonasal malignancies: an overview with focus on the current WHO classification and recently described provisional entities
Virchows Archiv (2024)
-
Molecular pathology in diagnosis and prognostication of head and neck tumors
Virchows Archiv (2024)
-
Top 10 Histological Mimics of Neuroendocrine Carcinoma You Should Not Miss in the Head and Neck
Head and Neck Pathology (2023)
-
Histopathologic Diagnosis of Sinonasal Tumors: Challenges and the Importance of Establishing the Correct Diagnosis
Current Otorhinolaryngology Reports (2023)