Abstract
Anastomosing hemangiomas are recently described benign vascular lesions that occur chiefly in the genitourinary tract and paravertebral soft tissues. Owing to their rarity and unusual cytoarchitectural features, anastomosing hemangiomas are frequently confused with low-grade angiosarcomas. The specific genetic alterations underlying these lesions are currently unknown. We performed capture-based next-generation DNA sequencing analysis on 13 anastomosing hemangiomas and identified frequent somatic mutations in the heterotrimeric G-protein alpha-subunit, GNAQ. Nine of 13 cases (69%) harbored a somatic mutation at GNAQ codon 209, a known hotspot that is commonly mutated in uveal melanoma and blue nevi, as well as various congenital vascular proliferations. No other pathogenic or likely pathogenic mutations were identified in these genetically simple lesions. The finding of a recurrent driver mutation in the G-protein signal transduction pathway provides strong evidence that anastomosing hemangiomas are indeed clonal vascular neoplasms.
Similar content being viewed by others
Main
Anastomosing hemangioma is a recently described vascular lesion that occurs predominantly in older adults. Initially thought to be unique to the genitourinary system, they have since been reported to occur in the adrenal gland, ovary, gastrointestinal tract, paravertebral soft tissue, and liver, with fewer than 80 cases reported in the literature.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 Histologically, anastomosing hemangiomas consist of a non-lobular proliferation of tightly packed, capillary-sized vascular channels, often with fibrin thrombi, so-called ‘hobnail’ endothelial cells, intracytoplasmic eosinophilic globules, and extramedullary hematopoiesis. Although the non-lobular growth pattern and prominent endothelial cells of anastomosing hemangioma often prompt concern for low-grade angiosarcoma by pathologists unfamiliar with this entity, all available evidence strongly supports the benign nature of these lesions.1, 3, 5, 8, 9
Recent data have implicated specific genetic alterations in the pathogenesis of benign vascular neoplasms. Among vascular entities, it has been reported that IDH1 mutations are highly specific for spindle cell hemangiomas,16 whereas congenital hemangiomas and capillary malformations of Sturge–Weber syndrome have been found to harbor activating GNAQ mutations.17, 18, 19 The molecular pathogenesis of anastomosing hemangiomas has not been described. In the present study, we examined the genomic features of anastomosing hemangiomas using capture-based next-generation sequencing of ~500 cancer-related genes with the goal of identifying mutations to further characterize these lesions and differentiate them from similar vascular tumors.
Materials and methods
Study Population
This study was approved by the institutional review boards of the University of California San Francisco and Mayo Clinic, Rochester, MN, USA. Cases were identified by examining excision specimens diagnosed as hemangiomas, vascular lesions, or angiosarcomas from the archives of the Department of Pathology of UCSF, spanning years from 1998 to 2016. Over 300 vascular lesions were reviewed. Lesions with predominantly capillary-sized vessels were identified and further evaluated for the presence or absence of features of anastomosing hemangiomas, yielding 16 lesions with at least one histologic feature. Four additional cases previously diagnosed as ‘anastomosing hemangioma’ were obtained from the consultation archives of one of the authors (ALF). These 20 cases were then re-reviewed in a blinded fashion by two of the authors (AEH and ALF). Thirteen cases were felt by both observers to show classical morphological features of anastomosing hemangioma1, 8 and comprise the final study population.
Capture-Based Next-Generation DNA Sequencing
Matched normal and tumor tissues were selected from 10 anastomosing hemangiomas. In three cases, no normal tissue was available and only tumor tissue was analyzed. Capture-based next-generation sequencing was performed at the UCSF Clinical Cancer Genomics Laboratory, using an assay (UCSF500 panel) that targets the coding regions of ~500 cancer-related genes, select introns from approximately 40 genes, and the TERT promoter with a total sequencing footprint of 2.8 Mb. Sequencing libraries were prepared from genomic DNA extracted from punch biopsies or macrodissected unstained sections from formalin-fixed paraffin-embedded tissue. Target enrichment was performed by hybrid capture using a custom oligonucleotide library. Sequencing was performed on a HiSeq 2500 (Illumina, San Diego, CA). Duplicate sequencing reads were removed computationally to allow for accurate allele frequency determination and copy number calling. The analysis was based on the human reference sequence UCSC build hg19 (NCBI build 37), using the following software packages: BWA: 0.7.10-r789, Samtools: 1.1 (using htslib 1.1), Picard tools: 1.97 (1504), GATK: 2014.4-3.3.0-0-ga3711, CNVkit: 0.3.3, Pindel: 0.2.5a7, SATK: 2013.1-10- gd6fa6c3, Annovar: v2015Mar22, Freebayes: 0.9.20 and Delly: 0.5.9.20, 21, 22, 23, 24, 25, 26, 27, 28, 29 Only insertions/deletions (indels) up to 100 bp in length were included in the mutational analysis. Somatic single-nucleotide variants and indels were visualized and verified using Integrated Genome Viewer. Genome-wide copy number analysis based on on-target and off-target reads was performed by CNVkit and Nexus Copy Number (Biodiscovery, Hawthorne, CA).
Results
Clinical Features
The clinical features of the 13 cases of anastomosing hemangiomas analyzed in this study are shown in Table 1. Three of these cases have been previously reported.1 All cases occurred in adults, with age at resection ranging from 39 to 79 years (mean, 60 years). They were primarily small lesions, with an average size of 2.8 cm (range 0.5–8 cm). Four were associated with the genitourinary tract, four were paraspinal, and three were gynecological. The majority of cases were discovered incidentally at the time of work up for other disease processes. Two patients presented with neurological symptoms owing to spinal cord compression. All patients with available data were without recurrence at the time of last clinical follow-up (average follow-up 32 months, range 1–107 months).
Histologic Features
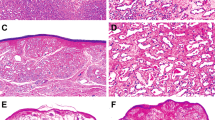
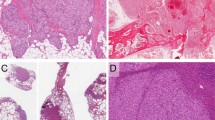
The histologic features of the 13 cases are summarized in Table 2. All cases were composed of capillary-sized vascular channels with rare to absent mitoses and mild to no cytologic atypia (Figure 1). All cases had small fibrin thrombi (13/13, 100%), and the majority of cases had focal to abundant intracytoplasmic eosinophilic globules (12/13, 92%) and extramedullary hematopoiesis (7/13, 54%) (Figures 2a and c). Some cases showed prominent hobnailing of endothelial cells (5/13, 38%) (Figure 2d). In cases where the border was able to be evaluated, the lesions were relatively well-circumscribed, but two cases showed an irregular interface with the surrounding fibroadipose tissue (Figure 2e). Six of 13 (46%) cases had at least focal areas of degenerative changes or sclerosis (Figure 2f). Many cases showed scattered associated mast cells and foamy histiocytes.
Histologic features of anastomosing hemangiomas. Small fibrin thrombi were seen in all cases (a). The majority of cases showed focal to abundant intracytoplasmic eosinophilic globules (b) and extramedullary hematopoiesis, shown here with erythroid precursors (c). Prominent hobnailing of endothelial cells were noted in many cases (d). Two cases showed an irregular interface with the surrounding fibroadipose tissue (e). Many cases showed at least focal areas of degeneration or sclerosis (f).
Genomic Features
All 13 cases were tested using the targeted next-generation sequencing panel. Nine of 13 cases (69%) showed a GNAQ hotspot mutation at codon 209. Eight of nine had GNAQ p.Q209H mutations and the other one of nine had a GNAQ p.Q209L mutation (Table 2 and supplementary Table 1). No cases had any other pathogenic or likely pathogenic mutations. Four cases had no identifiable driver mutations. No cases had detectable copy number alterations, including chromosomal gains or losses as well as focal amplifications or deletions.
Discussion
Hemangiomas are common soft tissue lesions, the majority of which arise in the skin and subcutis. They include lobular capillary hemangioma (pyogenic granuloma), spindle cell hemangioma, epithelioid hemangioma (angiolymphoid hyperplasia with eosinophilia), hobnail hemangioma (targetoid hemosiderotic hemangioma), sinusoidal hemangioma, tufted angioma/hemangioma, and glomeruloid hemangioma. In children, common vascular lesions also include infantile hemangioma, noninvoluting congenital hemangioma, rapidly involuting congenital hemangioma, and partially involuting capillary hemangioma. Many of these have historically been considered reactive lesions, but more recent genetic studies have demonstrated clonal pathogenic mutations in some, suggesting a neoplastic nature.16, 17, 18, 19
Anastomosing hemangiomas differ from most other true hemangiomas inasmuch as they are typically deeply (or viscerally) situated and occur in adult patients. The present study is the first to examine their genetic alterations. We have identified a hotspot mutation in GNAQ codon 209 in 9 of 13 anastomosing hemangiomas (69%, Table 2). The presence of a recurrent mutation in this otherwise genetically simple lesion implicates GNAQ as a driver in its pathogenesis and confirms its clonal nature. No other pathogenic or likely pathogenic mutations were identified, including recurrent mutations described in angiosarcomas such as in PTPRB and PLCG1.30 Moreover, as mutations in GNAQ have not been reported in angiosarcomas, these results strongly suggest that anastomosing hemangiomas are not genetically related to angiosarcomas.
GNAQ encodes G-protein subunit alpha q, which together with its paralogues GNA11, GNA14, and GNA15 comprise the alpha q subfamily of G proteins, which are involved in mediating downstream signals between G-protein coupled receptors and downstream targets. Mutations in GNAQ at codon 209, alter a region within the catalytic GTPase domain, resulting in constitutive activity. Somatic mutations in GNAQ were first described in melanocytic lesions, and are present in ~50% of primary uveal melanomas and 80% of blue nevi, with either mutations at Q209 or R183.31 Similarly, such lesions show mutually exclusive GNA11 mutations at lower frequencies.32 Overexpression of GNAQ Q209L in primary human melanocytes increases signaling through the MAPK pathway.31 In this study, GNA11 hotspots also had greater than 400 × coverage and no mutations were seen in any of the cases. GNA14 and GNA15 were not assayed in this study; thus it is possible the cases lacking mutations in GNAQ could harbor mutations in either of these other genes or in other members of the pathway.
Interestingly, alterations in this family have been reported in other benign vascular tumors, in particular congenital vascular lesions. Recurrent mutations at codon 209 of GNAQ and GNA11 have been found in the majority of congenital hemangiomas (12/16 cases), using massively parallel mRNA sequencing.17 Specifically, 8/16 cases demonstrated GNAQ Q209L/P/H mutations, whereas 4/16 cases demonstrated mutually exclusive GNA11 Q209L mutations. GNA14 Q205L mutations have also been reported in one case each of lobular capillary hemangioma, tufted angioma, and kaposiform hemangioendothelioma using whole-exome and targeted sequencing, with GNA11 R183C mutations in two cases of lobular capillary hemangioma.19 Expression of mutated GNA11 or GNA14 in primary human umbilical vein endothelial cells has been shown to activate the MAP kinase pathway and render cells growth-factor independent.19
Capillary malformations in Sturge–Weber syndrome and nonsyndromic port-wine stains have also been shown to demonstrate a somatic GNAQ mutation at R183Q/L/G.18, 33 Unlike true capillary hemangiomas, these lesions are thought to be malformative, and contain abnormally configured thick-walled vessels in addition to capillaries. Among hematopoietic, perivascular, and stromal cells, fluorescence-activated cell sorting studies have identified endothelial cells as the population enriched for GNAQ mutation in these lesions.33
A vascular liver lesion termed hepatic small vessel neoplasm has recently been described which demonstrates some morphological and clinical overlap with anastomosing hemangioma.34 This entity was defined as an infiltrative vasoformative neoplasm composed of small vessels without diagnostic features of cavernous hemangioma or angiosarcoma.34 Interestingly, 2/3 cases of hepatic small vessel neoplasm showed GNAQ Q209H mutations and one of these cases also had a hotspot PIK3CA mutation. In contrast, the published series of four hepatic anastomosing hemangiomas were sharply demarcated from surrounding liver and 2/4 had intermixed features of cavernous hemangioma.9 Given morphological and molecular similarities between hepatic small vessel neoplasm and anastomosing hemangiomas from other sites, it may be possible that hepatic small vessel neoplasm represents an anastomosing hemangioma with more infiltrative growth than is typical in other sites, but sequencing of additional cases is needed.
In summary, we have shown for the first time recurrent GNAQ mutations in anastomosing hemangiomas. Although other capillary hemangiomas, in particular congenital hemangiomas, also have GNAQ mutations, the clinical setting (namely patient age and location) makes anastomosing hemangiomas unique within this group. In addition, GNAQ mutations are not found in angiosarcoma, and there may be a role for identification of GNAQ mutations in the distinction between anastomosing hemangiomas and low-grade angiosarcoma, although study of a larger number of cases is required. Taken together, the finding of driver mutations in G-protein coupled signal transduction confirms the clonal nature of anastomosing hemangiomas and supports a role for this pathway in the pathogenesis of benign vascular tumors, distinct from the pathogenesis of malignant vascular tumors.
References
John I, Folpe AL . Anastomosing hemangiomas arising in unusual locations: a clinicopathologic study of 17 soft tissue cases showing a predilection for the paraspinal region. Am J Surg Pathol 2016; 40: 1084–1089.
Kryvenko ON, Gupta NS, Meier FA et al, Anastomosing hemangioma of the genitourinary system: eight cases in the kidney and ovary with immunohistochemical and ultrastructural analysis. Am J Clin Pathol 2011; 136: 450–457.
Kryvenko ON, Epstein JI . Testicular hemangioma: a series of 8 cases. Am J Surg Pathol 2013; 37: 860–866.
Kryvenko ON, Haley SL, Smith SC et al, Haemangiomas in kidneys with end-stage renal disease: a novel clinicopathological association. Histopathology 2014; 65: 309–318.
Brown JG, Folpe AL, Rao P et al, Primary vascular tumors and tumor-like lesions of the kidney: a clinicopathologic analysis of 25 cases. Am J Surg Pathol 2010; 34: 942–949.
Heidegger I, Pichler R, Schafer G et al, Long-term follow up of renal anastomosing hemangioma mimicking renal angiosarcoma. Int J Urol 2014; 21: 836–838.
Jin LU, Liu J, Li Y et al, Anastomosing hemangioma: the first case report in the bladder. Mol Clin Oncol 2016; 4: 310–312.
Montgomery E, Epstein JI . Anastomosing hemangioma of the genitourinary tract: a lesion mimicking angiosarcoma. Am J Surg Pathol 2009; 33: 1364–1369.
Lin J, Bigge J, Ulbright TM et al, Anastomosing hemangioma of the liver and gastrointestinal tract: an unusual variant histologically mimicking angiosarcoma. Am J Surg Pathol 2013; 37: 1761–1765.
Mehta V, Ananthanarayanan V, Antic T et al, Primary benign vascular tumors and tumorlike lesions of the kidney: a clinicopathologic analysis of 15 cases. Virchows Arch 2012; 461: 669–676.
Metodiev D, Ivanova V, Omainikova B et al, [Ovarian anastomosing hemangioma with stromal luteinization: a case report]. Akush Ginekol (Sofiia) 2015; 54: 58–61.
Omiyale AO . Anastomosing hemangioma of the kidney: a literature review of a rare morphological variant of hemangioma. Ann Transl Med 2015; 3: 151.
Ross M, Polcari A, Picken M et al, Anastomosing hemangioma arising from the adrenal gland. Urology 2012; 80: e27–e28.
Tao LL, Dai Y, Yin W et al, A case report of a renal anastomosing hemangioma and a literature review: an unusual variant histologically mimicking angiosarcoma. Diagn Pathol 2014; 9: 159.
Tran TA, Pernicone P . Anastomosing hemangioma with fatty changes of the genitourinary tract: a lesion mimicking angiomyolipoma. Cent European J Urol 2012; 65: 40–42.
Kurek KC, Pansuriya TC, van Ruler MA et al, R132C IDH1 mutations are found in spindle cell hemangiomas and not in other vascular tumors or malformations. Am J Pathol 2013; 182: 1494–1500.
Ayturk UM, Couto JA, Hann S et al, Somatic activating mutations in GNAQ and GNA11 are associated with congenital hemangioma. Am J Hum Genet 2016; 98: 1271.
Shirley MD, Tang H, Gallione CJ et al, Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med 2013; 368: 1971–1979.
Lim YH, Bacchiocchi A, Qiu J et al, GNA14 somatic mutation causes congenital and sporadic vascular tumors by MAPK activation. Am J Hum Genet 2016; 99: 443–450.
Li H, Durbin R . Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010; 26: 589–595.
Yang H, Wang K . Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 2015; 10: 1556–1566.
Rausch T, Zichner T, Schlattl A et al, DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 2012; 28: i333–i339.
DePristo MA, Banks E . Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011; 43: 491–498.
Van der Auwera GA, Carneiro MO, Hartl C et al, From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 2013; 43: 11 0 1–11 033.
Ye K, Schulz MH, Long Q et al, Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-endshort reads. Bioinformatics 2009; 25: 2865–2871.
Picard: A set of tools (in Java) for working with next generation sequencing data in the BAM. Available fromhttp://broadinstitute.github.io/picard.
Garrison E, Marth G . Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 [q-bio.GN]. Available from:http://arxiv.org/abs/1207.3907.
McKenna A, Hanna M, Banks E et al, The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20: 1297–1303.
Li H, Handsaker B, Wysoker A et al, The sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079.
Behjati S, Tarpey PS, Sheldon H et al, Recurrent PTPRB and PLCG1 mutations in angiosarcoma. Nat Genet 2014; 46: 376–379.
Van Raamsdonk CD, Bezrookove V, Green G et al, Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009; 457: 599–602.
Van Raamsdonk CD, Griewank KG, Crosby MB et al, Mutations in GNA11 in uveal melanoma. N Engl J Med 2010; 363: 2191–2199.
Couto JA, Huang L, Vivero MP et al, Endothelial cells from capillary malformations are enriched for somatic GNAQ mutations. Plast Reconstr Surg 2016; 137: 77e–82e.
Gill RM, Buelow B, Mather C et al, Hepatic small vessel neoplasm, a rare infiltrative vascular neoplasm of uncertain malignant potential. Hum Pathol 2016; 54: 143–151.
Acknowledgements
This research was supported and funded by the UCSF Residents’ Teaching and Research Endowments from the UCSF Department of Pathology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Bean, G., Joseph, N., Gill, R. et al. Recurrent GNAQ mutations in anastomosing hemangiomas. Mod Pathol 30, 722–727 (2017). https://doi.org/10.1038/modpathol.2016.234
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.234
This article is cited by
-
A novel somatic mutation in GNAQ in a capillary malformation provides insight into molecular pathogenesis
Angiogenesis (2022)
-
Functional characterization of uveal melanoma oncogenes
Oncogene (2021)
-
Detection of cryptogenic malignancies from metagenomic whole genome sequencing of body fluids
Genome Medicine (2021)
-
GNA11 joins GNAQ and GNA14 as a recurrently mutated gene in anastomosing hemangioma
Virchows Archiv (2020)
-
What is new in endothelial neoplasia?
Virchows Archiv (2020)