Abstract
Xp11 translocation renal cell carcinomas are characterized by several different translocations involving the TFE3 gene. Tumors with different specific gene fusions may have different clinicopathological manifestations. Fewer than 10 renal cell carcinoma cases with NONO-TFE3 have been described. Here we examined eight additional cases of this rare tumor using clinicopathological, immunohistochemical, and molecular analyses. The male-to-female ratio of our study cohort was 1:1, and the median age was 30 years. The most distinctive feature of the tumors was that they exhibited glandular/tubular or papillary architecture that was lined with small-to-medium cuboidal to high columnar cells with indistinct cell borders and an abundantly clear or flocculent eosinophilic cytoplasm. The nuclei were oriented toward the luminal surface and were round and uniform in shape, which resulted in the appearance of secretory endometrioid subnuclear vacuolization. The distinct glandular/tubular or papillary architecture was often accompanied by sheets of epithelial cells that presented a biphasic pattern. Immunohistochemically, all eight cases demonstrated moderate (2+) or strong (3+) positive staining for TFE3, CD10, RCC marker, and PAX-8. None of the tumors were immunoreactive for CK7, Cathepsin K, Melan-A, HMB45, Ksp-cadherin, Vimentin, CA9, 34βE12 or CD117. NONO-TFE3 fusion transcripts were identified in six cases by RT-PCR. All eight cases showed equivocal split signals with a distance of nearly 2 signal diameters and sometimes had false-negative results. Furthermore, we developed a fluorescence in situ hybridization (FISH) assay to serve as an adjunct diagnostic tool for the detection of the NONO-TFE3 fusion gene and used this method to detect the fusion gene in all eight cases. Long-term follow-up (range, 10–102 months) was available for 7 patients. All 7 patients were alive with no evidence of recurrent disease or disease progression after their initial resection. This report adds to the known data regarding NONO-TFE3 renal cell carcinoma.
Similar content being viewed by others
Main
Renal cell carcinomas associated with Xp11 translocations are rare and are characterized by several different translocations involving the TFE3 gene. In these tumors, the TFE3 gene is fused by translocation to one of several other genes, including ASPL, PRCC, NONO (p54nrb), CLTC, SFPQ1, LUC7L3, KHSRP, PARP14, DVL2, RBM10, and unknown genes on chromosomes 10.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 Tumors with different specific gene fusions may have different clinical manifestations and morphological features.7 The first description of a NONO-TFE3 gene fusion came from a UOK109 papillary renal cell carcinoma cell line and was reported by Clark et al.1 Recently, Argani et al12 provided the first morphological description of NONO-TFE3 renal cell carcinoma, which was based on five cases. To the best of our knowledge, fewer than 10 cases of NONO-TFE3 renal cell carcinoma have been described in the literature to date.1, 12, 14, 15 Thus, the clinical manifestations and morphological features of NONO-TFE3 renal cell carcinoma must be further elucidated.
The detection of strong nuclear TFE3 immunoreactivity using an antibody to the C-terminal portion of TFE3 is currently the most commonly used diagnostic technique for Xp11 translocation renal cell carcinoma.16, 17 However, false-positive and false-negative results are quite frequent due to differences in fixation times, technical methods and scoring systems.7 TFE3 break-apart fluorescence in situ hybridization (FISH) assays on formalin-fixed paraffin-embedded tissue sections is currently the gold standard for identifying TFE3 rearrangements and often results in large-space split signals from a translocation.7 However, when TFE3 rearrangement results from an inversion of chromosome X, as in the NONO-TFE3 fusion, the split signals are more subtle.
In the current work, we performed a retrospective study of eight patients with NONO-TFE3 renal cell carcinoma and described the specific tumor morphology as well as the patients’ long-term survival. We also discussed a potential diagnostic pitfall in detecting NONO-TFE3 gene rearrangement using TFE3 break-apart FISH that can easily produce false-negative and equivocal results. Additional FISH assays may provide further confirmation of the presence of the NONO-TFE3 fusion gene.12, 13, 18
Materials and methods
Case Selection
Of the eight cases included in this study, one case was from the State Key Laboratory of Cancer Biology, Department of Pathology, Xi Jing Hospital, Fourth Military Medical University, Xi'an, China; one case was from the Department of Pathology, West China Hospital, West China Medical School, Sichuan University, Chengdu, China; one case was from the Department of Pathology, Fudan University Shanghai Cancer Center, Shanghai, China; and five cases were from the archives of the Department of Pathology at Nanjing Jinling Hospital, Nanjing University School of Medicine. Clinicopathological characteristics, treatments, and follow-up data were recorded.
For each case, the pathology report and all tissue sections stained with hematoxylin and eosin (mean, 7.1 slides; median, 7 slides; range, 4–13 slides) were reviewed independently by experienced pathologists (RQ and XQY). The blocks containing the largest proportion of tumor tissue for each case were selected for immunostaining.
Immunohistochemistry
Tumor tissues were fixed in 10% formalin and embedded in paraffin. Sections that were 3-μm thick were immunohistochemically stained using antibodies against the following proteins: TFE3 (SC-5958, 1:300; Santa Cruz, Santa Cruz, CA, USA), cathepsin K (3F9, 1:300; Abcam, Cambridge, UK), HMB45 (1:500; Dako, Glostrup, Denmark), Melan-A (A103/M2-72, 1:100; NeoMarkers, Fremont, CA, USA), CD10 (56C6, 1:100; Novocastra, Milton Keynes, UK), vimentin (V9, 1:200; Zymed, Grand Island, NY, USA), CK7 (OV-TL12/30, 1:300; Zymed), CD117 (Polyclonal, 1:100; Dako), Ksp-cadherin (4H6/F9, 1:200; Zymed), PAX8 (4H7B3, 1:100; ProteinTech Group, Rosemont, IL, USA), RCC marker (PN-15, pre-diluted; MaxVision, Madison, AL, USA), carbonic anhydrase IX (CA9) (ab1508, 1:1000; Abcam) and cytokeratin 34βE12 (pre-diluted, Ventana, Roche, Tucson, AZ, USA).
Immunoreactions were performed using labeled streptavidin-biotin and overnight incubation as previously described. Then, 3,3’-diaminobenzidine was used for visualization. Immunoreactivity was evaluated in a semiquantitative manner to assess both staining intensity and the percentage of immunopositive cells as described previously.10, 19 For all antibodies, the resulting score was calculated by multiplying the staining intensity (0=no staining, 1=mild staining, 2=moderate staining and 3=strong staining) by the percentage of immunoreactive tumor cells (0–100). The immunostaining result was considered 0 or negative when the score was <25; it was considered 1+ or weak when the score was 26–100; it was considered 2+ or moderate when the score was 101–200; and it was considered 3+ or strong when the score was 201–300.
Detection of the NONO-TFE3 Fusion Gene via RT-PCR
An RT-PCR assay was performed as described by Clark et al and Chang et al20 to detect major TFE3-related fusion genes, including t(X;1) (p11.2;p34) SFPQ1-TFE3, t(X;17)(p11.2;q23) CLTC-TFE3, inv(X)(p11.2;q12) NONO-TFE3, t(X;1)(p11.2;q21) PRCC-TFE3, and t(X;17)(p11.2;q25.3) ASPL-TFE3.1, 20
For the NONO-TFE3 fusion gene, new primer pairs were designed to detect fusion gene transcripts. These primers are listed in Supplementary Table S1.
For sequence analysis, the PCR products were purified using a Wizard PCR Preps Purification System (Promega, Madison, WI, USA). Sequencing was performed using Big Dye Terminator and ABI Basecaller (Applied Biosystems, Grand Island, NY, USA).
FISH Probe Design and Development
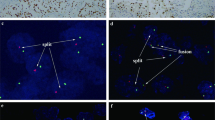
Bacterial artificial chromosome clones were selected using the 'CloneCentral human bacterial artificial chromosome Clone Locator' from EmpireGenomics (http://www.empiregenomics.com/CloneCentral/gene_search) as previously described for split hybridization experiments.7, 10, 13, 21 For the NONO-TFE3 fusion assay, the bacterial artificial chromosome clone RP11-934E1, located centromeric to the NONO gene locus, was labeled with 5-fluorescein-dUTP. The bacterial artificial chromosome clone RP11-416B14, located telomeric to the TFE3 gene locus, was labeled with 5-ROX-dUTP (Figure 1a).
FISH
One slide stained with hematoxylin and eosin from each block was examined to identify areas containing tumor cell clusters for cell counting. Tissue sections that were 3-μm thick were prepared from buffered formalin-fixed paraffin-embedded tissue blocks. Tumor tissues on the slides were deparaffinized and subjected to heat pretreatment (pressure cooking for 10 min at full pressure) in distilled water and then digested by incubation with 0.25% pepsin (Sigma, Taufkirchen, Germany) and 0.01 M HCl for 15 min at 37 °C. After rinsing twice in 2xSSC for 5 min, the tissues were dehydrated by immersing the slides in 70%, 85%, and 100% ethanol for 1 min each at room temperature and then air-dried. The probes were diluted in tDenHyb 2 (Insitus, Albuquerque, NM, USA) at a ratio of 1:25. The slides containing the tissue DNA probes (10 μl per slide) were co-denatured in an in situ thermocycler (System 1000, Perkin Elmer, Waltham, Massachusetts, USA) at 83 °C for 12 min, annealed at 37 °C, and hybridized in a humidified chamber at 37 °C overnight. After post-hybridization washing in 0.4 × SSC (70 °C for 2 min) and 2 × SSC (room temperature for 2 min), a coverslip was added to the slides with 10 ml of 4,6-diamino-2-phenylindole for counterstaining.
FISH Evaluation
The method of analysis used for FISH evaluation has been partially described previously.7, 10, 22 The first step involved using a split probe assay to identify TFE3 gene rearrangement. A fused or closely approximated green-red signal pattern was interpreted as a normal result, whereas a split signal pattern indicated the presence of a TFE3 translocation.
The second step involved using a fusion probe assay with a probe telomeric to TFE3 (5-ROX-dUTP, red) and another probe centromeric to NONO (5-fluorescein-dUTP, green). A colocalized signal represented a fusion between NONO and TFE3 (Figure 1b). Signals were considered to be split when green and red signals were separated by a distance equal to or greater than the two signals’ diameters. For each case, a minimum of 100 tumor nuclei was examined for probe signals via fluorescence microscopy at 1000x magnification. To avoid false-positive interpretations resulting from nuclear truncation, only non-overlapping tumor nuclei were evaluated. Based on the generally accepted guidelines used by all other commercially available break-apart FISH assays and the TFE3 break-apart FISH assays developed here, a positive result was reported when more than 10% of the nuclei in a tumor tissue sample displayed evidence of a TFE3 gene rearrangement or the NONO-TFE3 fusion.7, 10, 21
As negative controls, four clear cell renal cell carcinomas, two papillary renal cell carcinomas, three ASPL-TFE3 renal cell carcinomas, two PRCC-TFE3 renal cell carcinomas, and a panel of non-neoplastic renal tissues were also evaluated.
Results
Patients
The clinicopathological features of the eight patients diagnosed with NONO-TFE3 renal cell carcinoma are shown in Table 1. The patients ranged in age from 23 to 61 years (mean, 32.5 years; median, 30 years). The male-to-female ratio was 1:1. According to the 2010 American Joint Committee on Cancer TNM staging system, five patients presented with stage I disease and three presented with stage II disease. Follow-up data were available for seven patients (range, 10–102 months; mean, 51 months; median, 52 months). After the initial resection, all seven patients were alive with no evidence of disease.
Morphology
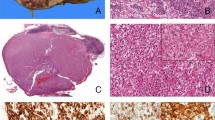
Morphologically, the most distinctive feature of the evaluated tumors was the presence of glandular/tubular or papillary architecture lined with small to medium cuboidal to high columnar cells with indistinct cell borders and abundant clear or flocculent eosinophilic cytoplasm (Figure 2a). The nuclei were oriented toward the luminal surface and were rounded and uniform in shape, resulting in the appearance of secretory endometrioid subnuclear vacuolization or a pattern that mimics clear cell papillary renal cell carcinoma, as previously described by Argani et al.12 The nucleoli were not prominent (WHO/ISUP grade 2; Figure 2b).
The morphologically distinct feature of NONO-TFE3 renal cell carcinoma is the presence of a biphasic pattern. (a) In case 6, the tumor had a predominantly glandular/tubular or papillary architecture lined by cuboidal to high columnar cells with abundantly clear or flocculent eosinophilic cytoplasm. (b) Nuclei were oriented toward the luminal surface and were round and uniform in shape, resulting in the appearance of secretory endometrioid subnuclear vacuolization. Focal psammoma bodies were also observed. (c) The proportion of sheets of epithelial cells and glandular/tubular or papillary architecture was equal in case 1. (d) High-power field microscopy of glands revealed the appearance of secretory endometrioid subnuclear vacuolization. (e) In case 2, the tumor was predominantly composed of solid sheets of epithelial cells. (f) Focal glands/tubules displaying the appearance of secretory endometrioid, which could be easily missed due to insufficient sampling.
Remarkably, this distinct glandular/tubular or papillary architecture was often accompanied by sheets of epithelial cells and therefore presented a biphasic pattern (Figures 2c–f). The proportion of glandular/tubular or papillary architecture observed for each case was variable and ranged from 5 to 90%. The architecture was predominantly glandular/tubular or papillary in three patients, predominantly solid sheets of epithelial cells in one patient, and equal for both morphologies in four patients. All eight patients exhibited psammoma bodies. Focal necrosis was observed in one patient. Hyaline degeneration of the stroma, hemorrhage, and hemosiderin were frequently observed.
Immunohistochemistry
All eight patients demonstrated moderate (2+) or strong (3+) positive staining for TFE3, CD10, RCC, and PAX-8. None of the patients were immunoreactive for CK7, Cathepsin K, Melan-A, HMB45, Ksp-cadherin, Vimentin, CA9, 34βE12, or CD117. The patients’ immunohistochemical profiles are summarized in Table 2.
Molecular Analysis
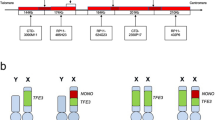
Adequate RNA was extracted from the formalin-fixed paraffin-embedded tissues from all eight patients and subjected to RT-PCR analysis. NONO-TFE3 fusion transcripts were identified in six patients (fusion transcript sequencing results are shown in Supplementary Figure S1). Sequencing of the PCR products revealed that the NONO-TFE3 fusion points were between exon 7 of NONO and exon 6 of TFE3 in 5 patients (Cases 1, 2, 4, 7, and 8) and between exon 9 of NONO and exon 5 of TFE3 in 1 patient (Case 5; Figure 3).
RT-PCR identified two NONO-TFE3 fusion gene patterns among six cases. One pattern had a fusion point between exon 7 of NONO and exon 6 of TFE3 (Cases 1, 2, 4, 7, and 8), while the other pattern had a fusion point between exon 9 of NONO and exon 5 of TFE3 (Case 5). A full color version of this figure is available at the Modern Pathology journal online.
FISH Analysis
Using the TFE3 break-apart FISH assay, all eight patients showed a high percentage (mean, 53%; range, 41–68%) of equivocal split signals with a distance of nearly two signal diameters and sometimes had false-negative results (Figures 4a and b).
Split (a and b) and fusion (c and d) FISH assays performed on tumors from female (Case 5) and male (Case 1) patients and showed positive results. Remarkably, the maintenance of glandular/tubular architecture could be visualized under a fluorescent microscope. A high percentage of the tumor cells that were arranged in a linear pattern inside the glands/tubules exhibited positive results, while the stroma cells in the surrounding fibrous and vascular septa were negative like the internal controls (b and d).
For the controls, three ASPL-TFE3 renal cell carcinomas and two PRCC-TFE3 renal cell carcinomas showed a positive result with a high percentage of widely split signals (mean, 59%; range, 47–72%), while the four clear cell renal cell carcinomas, two papillary renal cell carcinomas and non-neoplastic renal tissues were negative.
The fusion probe assay that used probes centromeric of NONO and telomeric of TFE3 showed a positive fusion signal. All 8 patients demonstrated a high percentage of cells with colocalized signals (mean, 59%; range, 51–75%; Figures 4c and d), whereas none of the 11 controls or the non-neoplastic renal tissues showed positive FISH results.
Discussion
Aside from ASPL, PRCC and SFPQ1, which are relatively common gene fusion partners associated with Xp11 translocation renal cell carcinoma, there are several exceedingly rare gene fusion partners, including NONO (p54nrb), CLTC, LUC7L3, KHSRP, PARP14, DVL2, and RBM10.1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 15 Many of these genes, such as CLTC, LUC7L3, KHSRP, PARP14, DVL2, and RBM10, have only been discussed in case reports.4, 8, 9, 11, 12 To the best of our knowledge, fewer than 10 NONO-TFE3 renal cell carcinomas have been described in the English literature to date.1, 12, 14, 15 In this study, eight patients were identified as having NONO-TFE3 renal cell carcinoma. The male-to-female ratio was 1:1, and there was no sex predominance. The patient ages ranged from 23 to 61 years (mean, 32.5 years; median, 30 years). All patients had a low pTNM stage (five with stage I, three with stage II). Long-term follow-up (range, 10–102 months; mean, 51 months; median, 52 months) was available for seven patients, all of whom were alive with no evidence of recurrent disease or disease progression after their initial resection. Information regarding the clinical characteristics of the NONO-TFE3 renal cell carcinomas remained unclear because of their rare incidence. In our study, NONO-TFE3 renal cell carcinoma appeared to be a relatively indolent tumor. In the patients with available treatment and prognostic information, there were no reports of death associated with NONO-TFE3 renal cell carcinoma.1, 12, 14, 15 Although Argani et al12 reported metastasis in two of five patients, Kristyna et al15 reported recurrence in one of three patients, survival or deceased outcome was not reported. Our relatively small sample may not be indicative of the true outcome of these tumors. Therefore, their exact biological behavior should be determined through further investigations using a larger cohort.
Xp11 translocation renal cell carcinomas with different specific gene fusions may have slightly different clinical manifestations and morphological features.7, 16, 23 In a recent study conducted by Argani et al,12 four of five patients demonstrated nuclear palisading with subnuclear vacuoles, a pattern that mimics clear cell papillary renal cell carcinoma. In our series, we demonstrated that NONO-TFE3 renal cell carcinomas displayed a distinctive architecture that contains a biphasic pattern composed of sheets of epithelial cells and glandular/tubular or papillary architecture. These glandular/tubular or papillary regions were lined with cuboidal to high columnar cells with abundant clear or flocculent eosinophilic cytoplasm. The nuclei were oriented toward the luminal surface and were rounded and uniform in shape, resulting in the appearance of secretory endometrioid subnuclear vacuolization or a pattern that mimics clear cell papillary renal cell carcinoma, as previously described by Argani et al.12 However, in one case reported by Argani et al,12 nuclear palisading that resembled trabecular architecture was present, which led to an initial diagnosis of neuroendocrine neoplasm, and similar subnuclear vacuolization was also observed in SFPQ1-TFE3 renal cell carcinoma. Disregarding the RT-PCR results, the frequent distinctive architecture observed in the NONO-TFE3 renal cell carcinomas along with the positive TFE3 staining and the equivocal TFE3 FISH results allowed for the subtyping of Xp11 translocation renal cell carcinomas. Such subtyping is assumed to be helpful for detecting additional potential cases and for furthering clinicopathological investigation. However, we also note that the proportion of glandular/tubular or papillary architecture for each case was variable and ranged from 5 to 90%. Therefore, sufficient sampling is important to avoid overlooking distinctive morphologies.
We also compared the features of NONO-TFE3 renal cell carcinomas and clear cell papillary renal cell carcinomas based on the literature and our present study.12, 15, 24, 25 Table 3 summarizes the clinicopathological findings observed in our series of NONO-TFE3 renal cell carcinomas compared with clear cell papillary renal cell carcinomas. NONO-TFE3 renal cell carcinomas frequently present at a younger age and with high columnar cells with indistinct cell borders, WHO/ISUP grade 2 nuclei, a flocculent eosinophilic cytoplasm, psammoma bodies, hyaline degeneration of the stroma, hemorrhage, hemosiderin, and strong immunostaining of TFE3, which are features not typically observed in clear cell papillary renal cell carcinoma. clear cell papillary renal cell carcinomas usually present with glandular/tubular or papillary architecture with open and large lumens, intraluminal pale eosinophilic material, cuboidal to low columnar cells, a perfectly clear cytoplasm with relatively distinct cell borders, WHO/ISUP grade 1 nuclei in a linear arrangement, smooth muscle stroma, and immunostaining of CA9, 34βE12, and CK7 (Figure 5).
(a) NONO-TFE3 renal cell carcinomas frequently exhibited high columnar cells with indistinct cell borders, a flocculent eosinophilic cytoplasm, and WHO/ISUP grade 2 nuclei. Psammoma bodies were always observed in our series. (b) In NONO-TFE3 renal cell carcinomas, hyaline degeneration of the stroma, hemorrhage, and hemosiderin were usually observed. (c) clear cell papillary renal cell carcinomas usually demonstrated glandular/tubular or papillary architecture with open and large lumens, intraluminal pale eosinophilic material, cuboidal to low columnar cells, and perfectly clear cytoplasms with relatively distinct cell borders. The nuclei were mostly WHO/ISUP grade 1. (d) Fibrous and/or smooth muscle stroma in varying amounts was noted in the clear cell papillary renal cell carcinomas.
Most NONO-TFE3 renal cell carcinomas reported in references Argani et al12 and Kristyna et al15 and in the current study showed negative staining for cathepsin K (14/16, 87.5%). This contrasts with observations of mesenchymal tumors associated with the same gene fusion. Previous studies have demonstrated that renal carcinomas with SFPQ1-TFE3, NONO-TFE3, and DVL-TFE3 gene fusions are typically PAX8-positive and cathepsin K-negative, while mesenchymal tumors with the same gene fusion have the opposite immunophenotype.12, 13, 18, 26, 27 However, it has previously been shown that cathepsin K is frequently positive in a subset of Xp11 translocation-associated renal cell carcinomas. In the studies of Martignoni and Argani, 12 of 14 and 7 of 11 Xp11 renal cell carcinomas with the PRCC-TFE3 gene fusion showed a positive reaction with cathepsin K, but consistently negative in the ASPL-TFE3 renal cell carcinoma, suggesting that functional differences exist between the resulting fusion proteins.12, 26, 27
The NONO gene belongs to a conserved family of multifunctional nuclear factors termed DBHS (drosophila behavior human splicing) proteins, which also includes SFPQ1 and PSPC110.13, 28 Because it is involved in a variety of biological processes, including RNA splicing and editing, DNA unwinding and repair, gene transcription and stem cell differentiation, NONO is believed to play an important role in cancer.13, 28 Previous studies have reported highly overlapping functions between SFPQ1 and NONO, possibly resulting in the similar morphology of SFPQ1-TFE3 and NONO-TFE3 renal cell carcinomas.11, 12, 13
Various strategies have been developed to confirm the diagnosis of Xp11 translocation renal cell carcinoma.7, 16 Assessing nuclear TFE3 immunoreactivity using an antibody to the C-terminal portion of TFE3 is currently the most commonly used diagnostic technique for identifying Xp11 translocation renal cell carcinoma.7, 16, 17 However, false-positive and false-negative results are common due to differences in fixation times, technical methods and scoring system.7 RT-PCR is a highly specific technique, but it usually requires high-quality RNA (ie, fresh-frozen samples) and can only identify known TFE3 fusion variants.11, 29 This creates a major limitation because the spectrum of TFE3 fusion partners is constantly broadening.
Our group and previous researchers have noticed that NONO-TFE3 gene fusion can be identified based on TFE3 rearrangement features using FISH. Using this technique, an uncommon split with a fixed distance (~2 signal diameters) implies chromosome X inversion instead of translocation.12, 13 In this setting, a subtle TFE3 break-apart FISH pattern that does not clearly match the current recommended guidelines, which state that inter-spot spacing should be at least twice the spot diameter to consider a split signal as positive, could be missed by inexperienced observers. Furthermore, we developed a NONO-TFE3 fusion FISH assay to detect the NONO-TFE3 fusion gene in all eight of the included patients. The abnormally colocalized signal of the NONO-TFE3 fusion in the FISH assay appears to be more easily interpreted than the TFE3 split assay. However, Just et al recently identified a new recurrent inversion leading to RBM10-TFE3 renal cell carcinoma by formalin-fixed paraffin-embedded RNA-Seq.11 The RBM10-TFE3 fusion transcript resulting from a paracentric inversion would theoretically result in an even smaller split signal than the NONO-TFE3 fusion transcript and its TFE3 break-apart pattern was poorly illustrated by FISH.11 The referenced work showed that RNA-Seq can be used as a robust supplementary technique to detect fusion transcripts from formalin-fixed paraffin-embedded material.11 Taken together, the diagnosis of such tumors should be based not only on morphology but also on immunophenotype and molecular genetic findings, especially for tumors with unusual pathological manifestations.
In summary, we reported the details of eight patients with NONO-TFE3 renal cell carcinoma and described the specific morphology of the tumors. All of the patients showed good prognosis with long-term survival. It is too early to conclude that NONO-TFE3 renal cell carcinoma is a relatively indolent tumor, as our relatively small sample may have influenced our results and relatively short-term follow-up may not be indicative of the true outcomes of patients with these tumors. Therefore, further investigations of larger and more heterogeneous populations should be conducted to validate and extend our results. We also confirmed a potential diagnostic pitfall associated with using TFE3 break-apart FISH assays to detect NONO-TFE3 gene rearrangement that can easily lead to false-negative and equivocal results. We further developed a FISH assay to serve as an adjunct diagnostic tool for the detection of NONO-TFE3 fusion genes. This report adds to the known data regarding NONO-TFE3 renal cell carcinoma.
References
Clark J, Lu YJ, Sidhar SK et al, Fusion of splicing factor genes PSF and NonO (p54nrb) to the TFE3 gene in papillary renal cell carcinoma. Oncogene 1997; 15: 2233–2239.
Argani P, Hawkins A, Griffin CA et al, A distinctive pediatric renal neoplasm characterized by epithelioid morphology, basement membrane production, focal HMB45 immunoreactivity, and t(6;11)(p21.1;q12) chromosome translocation. Am J Pathol 2001; 158: 2089–2096.
Argani P, Antonescu CR, Couturier J et al, PRCC-TFE3 renal carcinomas: morphologic, immunohistochemical, ultrastructural, and molecular analysis of an entity associated with the t(X;1)(p11.2;q21). Am J Surg Pathol 2002; 26: 1553–1566.
Argani P, Lui MY, Couturier J et al, A novel CLTC-TFE3 gene fusion in pediatric renal adenocarcinoma with t(X;17)(p11.2;q23). Oncogene 2003; 22: 5374–5378.
Argani P, Olgac S, Tickoo SK et al, Xp11 translocation renal cell carcinoma in adults: expanded clinical, pathologic, and genetic spectrum. Am J Surg Pathol 2007; 31: 1149–1160.
Ross H, Argani P . Xp11 translocation renal cell carcinoma. Pathology 2010; 42: 369–373.
Rao Q, Williamson SR, Zhang S et al, TFE3 break-apart FISH has a higher sensitivity for Xp11.2 translocation-associated renal cell carcinoma compared with TFE3 or cathepsin K immunohistochemical staining alone: expanding the morphologic spectrum. Am J Surg Pathol 2013; 37: 804–815.
Malouf GG, Su X, Yao H et al, Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin Cancer Res 2014; 20: 4129–4140.
Huang W, Goldfischer M, Babyeva S et al, Identification of a novel PARP14-TFE3 gene fusion from 10-year-old FFPE tissue by RNA-seq. Genes Chromosomes Cancer 2015; 54: 500–505.
Rao Q, Shen Q, Xia QY et al, PSF/SFPQ is a very common gene fusion partner in TFE3 rearrangement-associated perivascular epithelioid cell tumors (PEComas) and melanotic Xp11 translocation renal cancers: clinicopathologic, immunohistochemical, and molecular characteristics suggesting classification as a distinct entity. Am J Surg Pathol 2015; 39: 1181–1196.
Just PA, Letourneur F, Pouliquen C et al, Identification by FFPE RNA-Seq of a new recurrent inversion leading to RBM10-TFE3 fusion in renal cell carcinoma with subtle TFE3 break-apart FISH pattern. Genes Chromosomes Cancer 2016; 55: 541–548.
Argani P, Zhong M, Reuter VE et al, TFE3-fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am J Surg Pathol 2016; 40: 723–737.
Wang XT, Xia QY, Ni H et al, Xp11 neoplasm with melanocytic differentiation of the prostate harboring the novel NONO-TFE3 gene fusion: report of a unique case expanding the gene fusion spectrum. Histopathology 2016; 69: 450–458.
Sato Y, Yoshizato T, Shiraishi Y et al, Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet 2013; 45: 860–867.
Pivovarcikova K, Grossmann P, Alaghehbandan R et al, letter to the editor, TFE3-fusion variant analysis defines specific clinicopathologic associations among Xp11 translocation cancers. Am J Surg Pathol 2016, e-pub ahead of print.
Argani P . MiT family translocation renal cell carcinoma. Semin Diagn Pathol 2015; 32: 103–113.
Argani P, Lal P, Hutchinson B et al, Aberrant nuclear immunoreactivity for TFE3 in neoplasms with TFE3 gene fusions: a sensitive and specific immunohistochemical assay. Am J Surg Pathol 2003; 27: 750–761.
Green WM, Yonescu R, Morsberger L et al, Utilization of a TFE3 break-apart FISH assay in a renal tumor consultation service. Am J Surg Pathol 2013; 37: 1150–1163.
Rao Q, Cheng L, Xia QY et al, Cathepsin K expression in a wide spectrum of perivascular epithelioid cell neoplasms (PEComas): a clinicopathological study emphasizing extrarenal PEComas. Histopathology 2013; 62: 642–650.
Chang IW, Huang HY, Sung MT . Melanotic Xp11 translocation renal cancer: a case with PSF-TFE3 gene fusion and up-regulation of melanogenetic transcripts. Am J Surg Pathol 2009; 33: 1894–1901.
Zhong M, De Angelo P, Osborne L et al, Dual-color, break-apart FISH assay on paraffin-embedded tissues as an adjunct to diagnosis of Xp11 translocation renal cell carcinoma and alveolar soft part sarcoma. Am J Surg Pathol 2010; 34: 757–766.
Rao Q, Liu B, Cheng L et al, Renal cell carcinomas with t(6;11)(p21;q12): a clinicopathologic study emphasizing unusual morphology, novel alpha-TFEB gene fusion point, immunobiomarkers, and ultrastructural features, as well as detection of the gene fusion by fluorescence in situ hybridization. Am J Surg Pathol 2012; 36: 1327–1338.
Argani P, Antonescu CR, Illei PB et al, Primary renal neoplasms with the ASPL-TFE3 gene fusion of alveolar soft part sarcoma: a distinctive tumor entity previously included among renal cell carcinomas of children and adolescents. Am J Surg Pathol 2001; 159: 179–192.
Srigley JR, Cheng L, Grignon DJ Clear cell papillary renal cell carcinoma. In: Moch H, Humphrey PA, Ulbright TM et al (eds). WHO Classification of Tumours of the Urinary System and Male Genital Organs. IARC Press: Lyon, France, 2016, pp 40–41.
Rao Q, Xia QY, Cheng L et al, Molecular genetics and immunohistochemistry characterization of uncommon and recently described renal cell carcinomas. Chin J Cancer Res 2016; 28: 29–49.
Martignoni G, Gobbo S, Camparo P et al, Differential expression of cathepsin K in neoplasms harboring TFE3 gene fusions. Mod Pathol 2011; 24: 1313–1319.
Martignoni G, Pea M, Gobbo S et al, Cathepsin-K immunoreactivity distinguishes MiTF/TFE family renal translocation carcinomas from other renal carcinomas. Mod Pathol 2009; 22: 1016–1022.
Medendorp K, van Groningen JJ, Schepens M et al, Molecular mechanisms underlying the MiT translocation subgroup of renal cell carcinomas. Cytogenet Genome Res 2007; 118: 157–165.
Macher-Goeppinger S, Roth W, Wagener N et al, Molecular heterogeneity of TFE3 activation in renal cell carcinomas. Mod Pathol 2012; 25: 308–315.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (81472391 to Qiu Rao and 81372743 to Xiao-jun Zhou).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Xia, Qy., Wang, Z., Chen, N. et al. Xp11.2 translocation renal cell carcinoma with NONO-TFE3 gene fusion: morphology, prognosis, and potential pitfall in detecting TFE3 gene rearrangement. Mod Pathol 30, 416–426 (2017). https://doi.org/10.1038/modpathol.2016.204
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.204
This article is cited by
-
Novel NONO::TFE3 fusion and ALK co-expression identified in a subset of cutaneous microcystic/reticular schwannoma
Virchows Archiv (2023)
-
TFE3 and TFEB-rearranged renal cell carcinomas: an immunohistochemical panel to differentiate from common renal cell neoplasms
Virchows Archiv (2022)
-
Nuclear translocation of ASPL-TFE3 fusion protein creates favorable metabolism by mediating autophagy in translocation renal cell carcinoma
Oncogene (2021)
-
The suitability of NONO-TFE3 dual-fusion FISH assay as a diagnostic tool for NONO-TFE3 renal cell carcinoma
Scientific Reports (2020)
-
NEAT1-TFE3 and KAT6A-TFE3 renal cell carcinomas, new members of MiT family translocation renal cell carcinoma
Modern Pathology (2019)