Abstract
The diagnosis of myelodysplastic syndrome (MDS) can be challenging, and may be facilitated by correlation with cytogenetic testing. Microarray analysis using comparative genomic hybridization and/or single-nucleotide polymorphism array can detect chromosomal abnormalities not seen by standard metaphase cytogenetics. We examined the ability of combined comparative genomic hybridization and single-nucleotide polymorphism analysis (hereafter referred to as ‘combined array’) to detect changes among 83 patients with unexplained cytopenias undergoing pathologic evaluation for MDS and compared results with 18 normal bone marrow controls. Thirty-seven patients (45%) were diagnosed with MDS, 12 patients (14%) were demonstrated to have ‘indeterminate dyspoiesis’ (insufficient for classification of MDS), 27 (33%) were essentially normal, and 7 patients (8%) had alternative pathologic diagnoses. Twenty-one MDS patients (57% of diagnoses) had effectively normal metaphase cytogenetics, but combined array showed that 5 of these (13% of MDS patients) harbored major cryptic chromosomal aberrations. Furthermore, nearly half of patients with ‘indeterminate dyspoiesis’ and 1 with normal morphology had clonal cytopenia(s) of undetermined significance by combined array analysis. Cryptic array findings among MDS patients and those with clonal cytopenias(s) included large-scale copy-neutral loss of heterozygosity (up to 118 Mb) and genomic deletion of loci implicated in MDS pathogenesis (eg, TET2 (4q22) and NUP98 (11p15)). By comparison, in MDS patients with abnormal metaphase cytogenetics, microarray mostly recapitulated findings seen by routine karyotype. Combined array analysis has considerable diagnostic yield in detecting cryptic chromosomal aberrations in MDS and in demonstrating aberrant clonal hematopoiesis in cytopenic patients with indeterminate morphologic dysplasia.
Similar content being viewed by others
Main
Myelodysplastic syndrome (MDS) is a spectrum of clonal stem cell hematological malignancies characterized by ineffective hematopoiesis. It manifests as peripheral cytopenias, dysplastic morphology of one or more hematopoietic cell type, and risk for progression to acute myeloid leukemia. MDS is currently classified according to strict morphologic, cytogenetic, and hematologic criteria.1 It may arise de novo, or as a secondary (eg, therapy-related) malignancy following exposure to chemotherapy or radiation.2 The importance of cytogenetics as a diagnostic tool is recognized by the World Health Organization classification system given that specific clonal cytogenetic abnormalities warrant a diagnosis of MDS (in the setting of refractory pancytopenia) even in the absence of morphologic dysplasia.
Approximately 40–50% of MDS patients do not have karyotype abnormalities detected using standard metaphase cytogenetics.3 Limitations include low resolution, a requirement for dividing cells, and the fact that balanced structural abnormalities (eg, translocation and inversions) are rare in MDS.3 Although recurrent chromosomal deletions/monosomies or gains are common (eg, partial/total deletions of chromosomes 5, 7, 13, 20, X/Y or trisomy 8), even these sometimes remain obscure.4, 5, 6 Fluorescence in situ hybridization (FISH) can overcome some limitations of metaphase cytogenetics, but it is typically used to detect only the most common chromosomal aberrations found in MDS and its utility is limited to those cases where adequate karyotype analysis is not available.7, 8 In contrast, microarray studies have been shown to identify frequent aberrations which are not detectable by metaphase cytogenetics alone. To date, multiple studies have shown the utility of performing microarray analysis to detect chromosomal aberrations in cases of known MDS and related hematologic malignancies.9 Cryptic amplifications/deletions (ie, copy number variation, or CNV) identified by comparative genomic hybridization are well-documented in MDS.10, 11, 12, 13, 14, 15, 16 Furthermore, copy-neutral loss-of-heterozygosity (LOH) (also known as uniparental disomy) detected by single-nucleotide polymorphism array has been shown to have prognostic significance in MDS.17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27 In samples with unsuccessful conventional cytogenetic results, or in specimens that are not amenable to routine cytogenetics, the detection rate of genomic aberrations by single-nucleotide polymorphism microarray is particularly useful.28, 29
The majority of microarray studies in MDS rely on annotated series of patients with known diagnoses.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29 To our knowledge, the diagnostic utility of microarray has not been evaluated in an unselected cohort of patients undergoing evaluation of peripheral cytopenias, nor has it been evaluated in patients who may be best categorized as having ‘idiopathic cytopenias of undetermined significance’. In this study we used combined array to detect copy-neutral LOH and cryptic CNV in the marrow of patients irrespective of diagnosis who were undergoing primary bone marrow evaluation for peripheral cytopenia(s) and/or clinical concern for MDS. We sought to determine the diagnostic utility of combined array analysis among patents with unexplained cytopenia(s) that have not been preselected with respect to diagnosis (including benign mimics of MDS), disease subtype, or conventional cytogenetic results. The term ‘clonal cytopenias of undetermined significance’ (CCUS) has been applied to cytopenic patients who do not meet criteria for MDS but have clonal hematopoiesis demonstrated by the detection of an acquired somatic mutation in an MDS-associated gene.30, 31 In this study, we extend the diagnosis of ‘CCUS’ to patients who lack routine morphologic or cytogenetic diagnostic criteria for MDS (or alternative diagnosis), but have aberrant clonal hematopoiesis demonstrated by combined array.
Materials and methods
Patient and Control Selection
The study cohort consisted of patients undergoing bone marrow biopsy and aspirate sampling for initial pathologic evaluation of peripheral cytopenia(s) and/or clinical suspicion of MDS from 2011–2013. Microarray analysis was performed in parallel with standard morphologic, flow cytometric, and cytogenetic evaluation. Selection of patients for inclusion in the study cohort was independent of any additional findings. Control cases were limited to normal marrow specimens selected from patients undergoing routine evaluation due to an alternative prior or concurrent malignancy, where no clinical suspicion of concurrent MDS was indicated at the time of analysis. Detailed clinical and pathologic features of all control cases are listed separately (Supplementary Table 1). Combined array analysis was performed on DNA from the same bone marrow aspirates samples submitted for cytogenetic analysis, with the exception of two cases (from patients 25 and 30) which relied on peripheral blood due to inadequate aspirate material at the time of evaluation. Test DNA was referenced against same-sex control DNA provided with Sure Tag DNA labeling kit (Agilent Technologies, Santa Clara, CA, USA). These studies were performed in compliance and with the approval of the Institutional Research Subjects Review Board.
Morphologic Evaluation
Morphologic assessment was performed on Wright-Giemsa stains bone marrow aspirate smears and hematoxylin and eosin stained formalin-fixed paraffin-embedded trephine core biopsy specimens. Morphologic dysplasia was evaluated based on diagnostic criteria according to the 2008 World Health Organization Classification of Tumours of Haematopoietic and Lymphoid Tissues.1 Cases of ‘indeterminate dyspoiesis’, in which the possibility of early or limited involvement by MDS could not be excluded on morphologic grounds alone, were identified when the frequency of dysplastic forms was below the 10% threshold for erythroid precursors, granulocytes, and megakaryocytes, or <15% ringed sideroblasts.
Cytogenetics and FISH
Cytogenetic analysis was carried out on blood or bone marrow aspirates using trypsin and Giemsa method. Karyotypes were described according to the International System for Human Cytogenetic Nomenclature (ISCN, 2009). FISH analyses were performed following the manufacturer’s instructions and standard protocols, as previously described.32 Normal cytogenetics was defined for the purposes of this study as an appropriate complement of 46 chromosomes and normal G-banding pattern in 20 metaphases, or normal G-banding with partial loss of Y chromosome in males over the age of 50 years. Also included in this group were cases which lacked definitive clonal abnormalities. These include cases with (1) normal karyotype and a limited number of metaphase spreads (<20) available for analysis, (2) isolated non-clonal karyotype aberrations (identified in a single cell and/or ruled-out by FISH as a true clonal abnormality), and (3) suspected constitutional abnormalities.
DNA Extraction
DNA was extracted from patient’s specimens (peripheral blood or bone marrow aspirates) using QIAamp DNA Blood Mini Kit (Cat # 51104; Qiagen, USA). NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, USA) was used for quantitation and evaluation of quality of the DNA.
Combined Comparative Genomic Hybridization and Single-Nucleotide Polymorphism Microarray
Microarray experiment was carried out on Agilent’s custom-designed whole-genome SurePrint G3 array 4 × 180 K, CCMC v2.0 (Agilent Technologies), plus cancer-targeted platform containing ~120,000 comparative genomic hybridization probes (60 mer) including 20,000 cancer-specific probes (1 probe/0.5–1 kb; minimum 1 probe/exon, maximum 200 probes/gene) covering over 500 cancer genes and >130 cancer-associated genomic regions and 60,000 single-nucleotide polymorphism probes which overlap with Alu1 and Rsa1 sites with minor allele frequency of >5%. Genotypes on this array are measured using one probe per single-nucleotide polymorphism, providing ~5–10 Mb resolutions for LOH detection across the entire genome. Briefly, patient and control DNA (1.0 μg) was digested with restriction enzymes, Alu1 and Rsa1, and enzymatically labeled with dyes Cyanine-5dUTP and Cyanine-3dUTP, respectively, using Sure Tag DNA labeling kit (Cat # 5190–3399) as per the manufacturer’s recommendations. The labeled DNA was hybridized as per the manufacturer’s recommendations at 65 °C for 40 h.
Data Analysis
Following washing, the slides were scanned in high-resolution scanner (Model # G2505C; Agilent Technologies) at 3-μm resolution. The output files were processed and analyzed by CytoGenomics software, v2.5 (Agilent Technologies), using an Aberration Detection Method 2 (ADM-2) algorithm for aberration analysis and visualization, with the threshold set at 6.0. To control small variations appearing in the data analysis and to get the clonal fraction (for mosaicism), we used an extra aberration filter, defining the minimum number of probes that should be present in an aberrant region as 5 (gain or loss), with the minimum absolute level of average log2 ratio of 0.15. The GC correction, diploid peak centralization, single-nucleotide polymorphism (confidence level 0.95) and LOH (threshold 6.0) parameters were included into the analysis. The cutoff for CNVs was set at 250 kb and confirmation was performed by FISH for all abnormal results.
Results
Patient and Control Characteristics
The clinicopathologic features of all patients included in the study are listed (Table 1). The study cohort (ie, patients undergoing evaluation for MDS) (n=83) had an average age of 70 years (median 70, range 26–91), and included 25 women and 58 men. Pathologic bone marrow evaluation showed 27 cases were morphologically unremarkable (ie, essentially normal findings), whereas 12 were noted to have ‘indeterminate dyspoiesis’ (mild dysplastic changes morphologically insufficient for a diagnosis of MDS). Thirty-seven cases were diagnostic of MDS (see Table 1 for specific subclassification). The seven remaining cases consisted of alternative hematologic malignancies or bone marrow disorders in the clinical/pathologic differential with MDS (see below and Supplementary Table 3).
The control cohort (n=18) had an average age of 62 years (median 61, range 49–79), which did not differ significantly from the study cohort (P=0.16, two-tailed unpaired t-test), and consisted of 4 women and 14 men. It included 8 patients in pathologic remission following prior chemotherapy treatment for an alternative hematologic malignancy, 2 patients with a history of carcinoma (only 1 of which received radiation therapy alone), and 8 additional untreated patients with negative staging bone marrows for newly diagnosed peripheral lymphoma. Average complete blood count data for the control group as a whole was within normal limits. Additional clinicopathologic features of the control group are detailed in Supplementary Table 1.
Evaluation for CNV and Copy-Neutral LOH in Controls
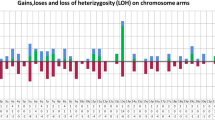
Among the control samples, no evidence of clinically significant CNV was present. Ten of 18 cases (56%), however, exhibited single-nucleotide polymorphism array findings that met study criteria for copy-neutral LOH (see the ‘Materials and Methods’ section; Table 2). With the exception of a single outlier case, these regions of copy-neutral LOH were limited to a range of 0.9–5.2 Mb, involved 1–4 separate chromosome arms per sample, and were distributed across 12 different chromosomes (1, 2, 5, 6, 8, 9, 10, 11, 12, 13, 15, and 18; Figure 1a and Supplementary Table 1). The outlier case (patient R) was a 79-year-old female with extensive copy-neutral LOH spanning 86 Mb on chromosome 4 (q12–q31.1), suggestive of clonal hematopoiesis in a morphologically normal bone marrow without resulting cytopenias(s) (see the ‘Discussion’ section for further evaluation).
Extent of copy-neutral LOH identified in normal controls versus morphologically normal bone marrow cases submitted for MDS evaluation. (a) Control samples, specimens A–R, with overlying black bars representing each individual contiguous segment of chromosomal copy-neutral LOH (chromosome arms listed above) for the corresponding case. Gray bar indicates one case classified as ‘clonal hematopoiesis of indeterminate potential’. (b) Morphologically normal bone marrow cases from the study cohort which exhibited normal cytogenetics (Group I), specimens 1 through 27, are displayed in a similar fashion. *indicate cases that had concurrent CNV detected by microarray comparative genomic hybridization. Additional karyotypic findings are indicated by underlined cases. Cases 20 and 21 demonstrated only partial loss of chromosome Y. #indicates cases with non-clonal karyotype aberrations that were otherwise non-informative (see Table 3). Dotted line indicates the 5 Mb threshold used for analysis throughout the study. For each case, bars are oriented in descending order, going from chromosomal region with longest to shortest segment of copy-neutral LOH. Corresponding chromosomal coordinates for segmental copy-neutral LOH are listed in Supplementary Table 1 and Table 3 for each group, respectively.
Identification of CNV and Copy-Neutral LOH in Samples Submitted for MDS Evaluation
Among the study cohort, 53 out of 83 samples (64%) had combined array findings, although the distribution of results was very different from controls (Table 2). Twenty-three of 83 cases (28%) demonstrated CNV (amplifications or deletions), whereas none were detected among the control group (P<0.01, Fisher’s exact test). Thirty cases had evidence of copy-neutral LOH alone, and many of these affected regions were substantially greater than those observed in the majority of controls (for examples see Figures 2 and 7). Despite individual cases with large segments of copy-neutral LOH in the study cohort, however, the overall burden in this group (measured as the average length of total copy-neutral LOH per genome per case) was not significantly different when compared with controls (mean of 17.0 Mb (±5.5 s.e.m.) versus 8.7 Mb (±4.7 s.e.m.), respectively; P=0.996, non-parametric Wilcoxon rank sum test). If comparison is limited to only those cases which exhibited segmental copy-neutral LOH substantially >5 Mb, more positive cases were detected among the study population (16 of 83 cases (19%) versus 1 of 17 (5.5%) among controls), but still the trend was not statistically significant (P=0.17, Fisher’s exact test).
Extent of copy-neutral LOH identified in cases with indeterminate dyspoiesis and normal cytogenetics. Specimens 28 through 39 (Group II), which exhibited insufficient morphologic dysplasia for MDS diagnosis and non-diagnostic cytogenetics. Case 34 (underlined) exhibited only loss of chromosome Y in a majority of metaphase spreads. Bars above each correspond to individual contiguous segment(s) of chromosomal copy-neutral LOH identified on separate chromosome arms, graphed in descending order from longest to shortest segment, with the involved chromosome arms listed above. Gray bars indicate cases classified as clonal cytopenia(s) of undetermined significance. *indicate cases which had concurrent CNV detected by microarray comparative genomic hybridization. Corresponding chromosomal coordinates for segmental copy-neutral LOH and CNV are listed in Table 4A.
Because the majority of cases in the study cohort (63 of 83, or 76%) exhibited essentially normal cytogenetics (see the ‘Materials and methods’ section for study criteria), the study cohort was subdivided into the following groups for further evaluation: Group I—morphologically normal bone marrow submitted for MDS evaluation with normal cytogenetics (Table 3); Group II—cases that showed ‘indeterminate dyspoiesis’ of uncertain clinical significance with normal cytogenetics (Table 4A); Group III—morphologic MDS with normal cytogenetics (Table 5); Group IV—morphologic MDS with abnormal cytogenetic findings (Supplementary Table 2); and Group V—alternative pathologic diagnoses (Supplementary Table 3).
Group I
Twenty-seven cases (32%) from the study cohort (average age 65 years, median 71) were morphologically within normal limits, with no evidence of malignancy or dysplasia and no definitive aberrant clonal cytogenetic abnormalities detected (Table 3). In this group single-nucleotide polymorphism analysis demonstrated only limited size copy-neutral LOH (<5 Mb) involving no more than three chromosomes (Figure 1b), similar to the majority of controls. Within these short segments of copy-neutral LOH, no clinically significant genes (ie, specifically implicated in MDS pathogenesis) were identified. Two patients (case numbers 20 and 21) demonstrated deletion of the entire chromosome Y, consistent with karyotype results. The only significant CNV in this group was from a single patient (case 10) involving an amplification of 255 kb at 6q27, including the locus encoding MLLT4 (AF6). This amplification was confirmed by FISH in 93% of cells, indicating clonal mosaicism and prompting classification as clonal cytopenia of undetermined significance (Table 3).
Group II
Twelve cases (14%) from the study cohort (average age 63 years, median 61) demonstrated only ‘indeterminate dyspoiesis’, insufficient for a morphologic diagnosis of MDS, and routine cytogenetics did not demonstrate a diagnostic abnormality. Notably, this subset of study patients had an age distribution that was highly similar to the controls (63±15 versus 62±10, P=0.87). Combined array analysis demonstrated a substantial number of these cases (5 of 12, 42% of Group II, or ~6% of total study cohort) harbored major chromosomal abnormalities indicative of clonal hematopoiesis and were therefore classified as clonal cytopenia(s) of undetermined significance (Table 4A and Figure 2). Specifically, four patient samples (case numbers 29, 30, 34, and 35) showed long segments of copy-neutral LOH (substantially >5 Mb, extending up to 118 Mb) on one or more chromosomes. One of these (case number 30) also showed an additional 2.3 Mb deletion at 12q24.31 that was subsequently confirmed by FISH in 44% of the interphase cells. In addition, CNV was found in isolation in a single sample (case number 32), which showed a 9 Mb deletion at 4q22.3-q24 (confirmed by FISH in 70% of interphase cells) and notably included TET2. The diagnostic combined array findings, laboratory data, and bone marrow results for several individual cases classified as clonal cytopenia(s) of undetermined significance are highlighted in Figures 3,4,5,6.
Case 29 (79-year-old male), submitted for indication of thrombocytopenia (platelets 122 × 103 cells/μl) and mild anemia (hemoglobin 11.8 mg/dl). Bone marrow aspirate exhibited only mild mekagaryocyte atypia but combined array demonstrated a 118 Mb segment of copy-neutral LOH on chromosome at 4q13.3-q35.2 (blue highlighted area) with an estimated clonal fraction of 90%. Follow-up demonstrated a mild absolute monocytosis (1.0 × 103 cells/μl), which persisted over a 30-month period, whereas platelet counts steadily declined (nadir of 89 × 103 cells/μl), but did not necessitate medical treatment during that time.
Case 30 (59-year-old female), submitted for severe longstanding thrombocytopenia (platelets 25 × 103 cells/μl) and mild anemia (hemoglobin 11.4 mg/dl). The aspirate and core biopsy showed no definitive evidence of dysplasia or increased blasts, but mild reticulin fibrosis was noted and rare circulating blasts were identified in the peripheral blood. Microarray identified several large segments of copy-neutral LOH on multiple chromosomes (including 36 Mb on 5q) and a single-genomic deletion of ~2.3 Mb at 12q24.31 (clonal fraction estimated at 65%). Deletion was confirmed by FISH in 63% of interphase cells. Bone marrow and peripheral blood microarray analysis performed 16 days apart showed nearly identical results. Follow-up at 18 months showed persistent thrombocytopenia and mildly worsening anemia without need for treatment. Subsequent bone marrow biopsies confirmed the presence of mild reticulin fibrosis, and mild myeloid dysplasia was noted on repeat biopsy 8 months after the original analysis. By 20 months post-analysis, bone marrow karyotype revealed a single aberrant cell with deletion of 5q in a background of a normal female karyotype (46,XX,del(5)(q13q33)[1]/46,XX[19]). Repeat combined array analysis at that time showed the same pattern of copy-neutral LOH and 12q24.31 deletion, in addition to a 2.6 Mb deletion at 11p11.2 and a 622 kb deletion at 11p13. Clonal fraction overall was estimated at 30–35%, and the first two deletions were confirmed by FISH in 39.5 and 33% of interphase cells, respectively. Treatment was limited to supportive care with occasional transfusions (peripheral counts declined to 7 × 103 platelets/μl, and hemoglobin of 10.1 mg/dl), and HLA-matching was performed for consideration of possible future bone marrow transplantation.
Case 32 (66-year-old male), submitted for pancytopenia (white blood cell count 2.1 × 103 cells/μl, hemoglobin 8.9 mg/dl, platelets 100 × 103 cells/μl), and showed very limited (<10%) morphologic irregularities among erythroid and megakaryocyte lineages in a background of overall myeloid predominant marrow. Metaphase cytogenetics was normal. Combined array demonstrated a 9-Mb deletion of 4q22.3-q24 that overlapped the TET2 locus (confirmed by FISH in 70% of cells). At follow-up 6 months later, the same deletion was confirmed at which time the clonal burden had increased to 94.5% of interphase cells (as determined by FISH). By 12 months post-analysis, peripheral counts were worsening, and the patient was unresponsive to erythropoietin therapy. Subsequent treatment included lenalidomide on clinical trial (four cycles), before transition to azacitidine with resulting significant improvement in peripheral blood counts.
Case 34 (88-year-old male) was a markedly hypocellular specimen with limited dyspoiesis, insufficient for diagnosis, but with loss of chromosome Y in 19 of 20 metaphase spreads and widespread chromosomal abnormalities detected as regions of copy-neutral LOH. The patient deceased within 18 months of analysis.
With a median follow-up of 28 months, the known clinical outcomes for patients with ‘indeterminate dyspoiesis’ are reported (Table 4B). Three of 3 patients with clonal cytopenia(s) of undetermined significance that underwent subsequent bone marrow/hematologic evaluation eventually met criteria for MDS or an MDS/MPN within 1–6 months. One patient was deceased within 18 months, and another had persistent macrocytic anemia, but was lost to follow-up after 7 months. Among those cases lacking evidence of clonal hematopoiesis by combined array, 4 of 7 received alternative benign hematologic diagnoses with overall improvement of peripheral blood counts in most cases, whereas 3 of 7 demonstrated persistent cytopenias and subsequently met criteria for MDS on repeat biopsy (within 7–9 months from analysis).
Group III
Twenty-one cases (25%) from the study cohort (average age 70 years, median 70) were diagnostic of MDS with essentially normal cytogenetics. These comprised 56% of new MDS diagnoses and encompassed the full spectrum of morphologic subtypes (Table 5). Four cases showed extensive copy-neutral LOH and CNV that went undetected by standard karyotype. Three patients (case numbers 45, 57, and 58) showed long-segment copy-neutral LOH (55, 59, and 38 Mb, respectively) on one chromosome arm each (Figure 7). One of these (number 58) had an additional 1.6 Mb deletion (11p15.5-p15.4) overlapping the locus for NUP98. A fifth patient (case number 41) demonstrated an isolated 322 kb deletion at 3q25.33, including at least three genes of unknown significance in MDS (see the ‘Discussion’ section). Both the 11p and 3q deletions were subsequently confirmed as clonal aberrations by FISH in 97 and 60% of cells, respectively. Together, these cases of MDS with cryptic chromosomal aberrations detected by combined array comprised nearly one-fifth of MDS with normal cytogenetics, 11% of total MDS, and ~6% of the entire study cohort in this analysis.
Extent of copy-neutral LOH identified in morphologic MDS cases with normal cytogenetics. Specimens 40 through 60 (Group III), with respective MDS diagnostic subclassification for each, and overlying black bars representing the extent of copy-neutral LOH detected on individual chromosome arms for each (as previously described in early figures). Cases 45 and 60 that demonstrated isolated loss of chromosome Y are also indicated. *indicate cases which had concurrent CNV detected by microarray comparative genomic hybridization. Corresponding chromosomal coordinates for segmental copy-neutral LOH and CNV are listed in Table 5.
Group IV
Sixteen cases (19%) were diagnostic of MDS (average age 72, median 73), with a variety of clonal cytogenetic abnormalities by metaphase cytogenetics (Supplementary Table 2). The majority of pathologic diagnoses were distinct morphologic subtypes of MDS, with two additional cases of therapy-related myeloid neoplasms. The spectrum of combined array findings in this group was broad, and in most cases correlated well with the available G-banding pattern. There were few examples of major chromosomal abnormalities detected by combined array (typically copy-neutral LOH) which were not apparent by metaphase cytogenetics (eg, case numbers 70, 74, and 75).
Group V
Seven other patients included in the study cohort (case numbers 77–83; average age 57 years, median 56) were diagnosed as having other clinical/pathologic entities well-recognized to induce peripheral cytopenias (including acute myeloid leukemia or high-grade borderline myeloid neoplasm). One case of borderline MDS/acute myeloid leukemia was found to contain a 2.4 Mb deletion at 9p24 (including the JAK2) that was otherwise associated with an apparent balanced t(8;9)(p21;p24) translocation (Supplementary Table 3). No other clearly significant findings were detected by karyotype or combined array among these cases.
Discussion
This study provides a number of important insights regarding diagnostic testing in MDS, particularly in cases with indeterminate findings, which are among the most diagnostically challenging and vexing cases for both pathologist and clinician alike. Whole-genome combined array analysis improves the ability to detect chromosomal abnormalities and uncovers potentially significant aberrations in a subset of patients with clinical and pathologic concern for MDS. In this study, 83 patients undergoing bone marrow aspiration/biopsy due to unexplained cytopenias were analyzed for clinically significant CNV and copy-neutral LOH alongside 18 controls. Positive-combined array findings among 10 control samples (56%) were largely limited to small stretches (<5 Mb) of copy-neutral LOH (see below for further discussion). This finding led us to conclude that segmental copy-neutral LOH >5 Mb could be used as threshold, above which results were interpreted as either suggestive or consistent with clonal hematopoiesis (depending on the extent and number of chromosomes involved). A total of 53 samples (64%) from the study cohort had positive combined array findings which included large-scale and widespread copy-neutral LOH as well as CNV. Thirty-seven samples (45%) in the study cohort were diagnostic of MDS, and another 6 cases (7% of study cohort) were classified as clonal cytopenia(s) of undetermined significance based on the combined array findings. The study cohort was organized in to 5 groups based on their morphological and cytogenetic/FISH findings. These groups (I–V) are discussed separately to highlight the significance of combined array in each.
Among cases submitted for MDS evaluation that were morphologically unremarkable and unrevealing cytogenetics (Group I), the overall pattern of combined array findings was similar to controls. Regions which met criteria for copy-neutral LOH did not significantly exceed 5 Mb and lacked any specific gene content information that was clinically actionable. Indeed, the overall pattern of results within this non-malignant subgroup so closely matches the controls that it warrants consideration as an internal control of aged patient’s with probable benign cytopenias, against which comparisons can be made with the remainder of the study cohort. Only one sample out of 27 (case number 10) exhibited a small-scale DNA duplication event (255 kb at 6q27), including gene AF6 (MLLT4), that was confirmed by FISH as clonal mosaicism and resulted in classification as clonal cytopenia of undetermined significance. The MLL-AF6 fusion oncogene which results from t(6;11)(q27;q23) translocation has been identified in wide range of hematologic malignancies, and it was recently found to be associated with poor outcome in childhood acute myeloid leukemia.33, 34 Overall, the findings among Group I patients indicate that in the absence of pathologic suspicion or concern for MDS, combined array does not offer significant diagnostic yield.
Group II consisted of 12 patients (14% of the study cohort) with indeterminate myelodysplasia, insufficient for morphologic diagnosis of MDS, and normal or otherwise non-definitive routine cytogenetics (including FISH). In this scenario, our results show that clinically significant CNV and copy-neutral LOH was detected in a substantial minority of cases (42%), warranting classification as clonal cytopenia(s) of undetermined significance. Currently, consensus guidelines do not exist for the type or pattern of comparative genomic hybridization or single-nucleotide polymorphism array findings that may be used to support or establish a diagnosis of MDS or clonal hematopoiesis. In the absence of such guidelines or objective criteria, the results for 4 patients from this study that were classified as clonal cytopenia(s) of undetermined significance (cases 29, 30, 32, and 34) are highlighted (Figures 3–6) to demonstrate the context in which combined array results impacted clinical assessment. Among the most subtle of these findings was an isolated 9 Mb deletion at 4q22.3-q24, overlapping the TET2 locus, which is among the most common mutated or altered genes in MDS.35, 36 The relatively high diagnostic yield of combined array in this group may be, at least in part, attributable to limited morphologic sampling in some cases, which precluded a definitive morphologic diagnosis. Regardless, these data support previous observations, where microarray analysis significantly improves the detection rate of clinically significant findings, particularly in cases with normal or unsuccessful conventional metaphase cytogenetics.20, 23, 28, 29
Group III patients received a morphologic diagnosis of MDS with ‘good’ or ‘very good’ cytogenetic risk category. A notable fraction of these cases (16–20%) had large-scale cryptic chromosomal aberrations found which otherwise went undetected by standard karyotype. Four patients (case numbers 46, 47, 59, and 60) showed segmental copy-neutral LOH above the set threshold (55, 8.7, 59, and 38 Mb, respectively). One of these (case number 60) had an additional 1.6-Mb deletion (11p15.5-p15.4) overlapping the locus for NUP98 which is associated with various hematopoietic malignancies as a proto-oncogene involved in numerous gene-fusion events.37 A fifth patient (case number 40) demonstrated an isolated 322-kb deletion at 3q25.33. The deleted region includes IFT80, SMC4, and C3orf80, and recently it was reported that SMC4 is strictly required for the proliferation and survival of NUP214-ABL1-positive T-cell acute lymphoblastic leukemia cells.38
The fourteen MDS cases from Group IV with clonal cytogenetic abnormalities by metaphase cytogenetics showed a wide spectrum of combined array findings that in most cases correlated well with the available G-banding pattern. There were few examples of major chromosomal abnormalities detected by combined array (typically copy-neutral LOH) which were not apparent by metaphase cytogenetics. For these few cases, the clinical significance of these microarray findings is uncertain given that they occurred in an already complex cytogenetic background and would not alter the patient’s prognosis based on the current risk-stratification scheme.
Group V included 6 cases that had other clinical/pathologic entities well-recognized to induce peripheral cytopenias (including acute myeloid leukemia and a high-grade borderline myeloid neoplasm) with only a single instance of a cryptic deletion being detected in an otherwise balanced translocation (for details see the ‘Results’ section).
Also worth highlighting here are the results from the control samples, in which single-nucleotide polymorphism probe hybridization-identified multiple small regions of the genome lacking heterozygosity. Such small regions of copy-neutral LOH (1–5 Mb) may be explained by autozygosity (the inheritance of the same ancestral loci from both parents), and are influenced by the degree of consanguinity within a population.25 Alternatively, the possibility that such findings represent small, acquired copy-neutral genomic aberrations among patients with history of cancer must also be considered. Similar evidence of clonal mosaicism in the peripheral blood, including copy-neutral LOH on the order of >2 Mb, has been associated with cancer in large-scale population studies.39, 40 All control patients in this study had a concurrent or prior diagnosis of malignancy, and approximately half of which had received prior chemotherapy treatment (a known predisposing risk factor for the development of MDS). Regardless, no significant differences were noted between those who received therapy, or between the group as a whole and the subset of study patients who also had morphologically normal marrow samples (Group I). The only exception to this pattern was the single outlier among the controls (patient R), a case best classified as ‘clonal hematopoiesis of indeterminate potential’, given that it is lacking both morphologic dysplasia and cytopenias.31 Additional clinical follow-up or diagnostic information was not available for this patient, but this case highlights the potential for combined array to detect clonal hematopoiesis in the elderly.
Limitations of this study include the lack of long-term follow-up for the majority of patients, selection of cases from a single institution, and an evaluation of how combined array findings might influence cytogenetic risk stratification among the diagnostic MDS cases. The use of bone marrow controls from patients with a history or current diagnosis of cancer is also a potential limitation. This population was intentionally chosen, however, to avoid the ambiguity of introducing presumed ‘benign’ syndromes (eg, aplastic anemia, ideopathic thromobocytopenic purpura, or macrocytic anemia) into this analysis, given that such conditions could be early or unrecognized manifestations of clonal hematopoiesis in a subset of patients. At the same time, this control group also provides a clinically relevant and ‘real-life’ basis for comparison, as evaluation of unexplained cytopenias and concern for MDS is routinely encountered in patients with history of cancer and chemo-radiation therapy. Against this backdrop, combined array analysis was able to discern a substantial fraction (42%) of cases with indeterminate morphologic findings were definitive clonal hematopoiesis.
Recent advances in our understanding of the molecular biology of MDS have identified a number of disease associated mutations.41, 42, 43 These somatically acquired mutations are relatively common in the general population among the aged, and are present irrespective of peripheral cytopenias.44, 45, 46 These mutations have been reported in 45–62% of patients with ICUS and some dysplasia.30 By comparison, 42% of cases with indeterminate dyspoiesis qualify as clonal cytopenia(s) of undetermined significance based on the combined array analysis in this study. Notably, our results found only a single case of clonal hematopoiesis among patients lacking dysplasia, whereas 17–20% of patient with ICUS and no dysplasia were found to be mutation positive.30 The question arises as to which modality provides a better indicator of clinically relevant clonal hematopoiesis. Among non-diagnostic samples from patients who subsequently developed a myeloid malignancy, the number of detectable point mutations is greater than in reported in healthy populations and predicts for worse outcome, whereas chromosomal aberrations detected by single-nucleotide polymorphism array are also common.47 Whether large-scale genomic changes, such as those described here (CNV>250 kb and/or copy-neutral LOH>5 Mb), represent a higher threshold and more specific measure of pathologic disorders, or conversely could represent a relatively late stage in the clonal evolutionary process, remains to be seen. In either case, care must be applied to avoid premature application of these advanced genomic techniques to the minimal criteria of malignant conditions.
Possible confounding factors in this type of analysis could be age-related clonality devoid of clinical significance, the transitory nature of clonal abnormalities in patients who received marrow modifying treatments (vitamins, chemotherapy, and so on), or constitutional genetic abnormalities. Given that all patients in this study cohort had cytopenias warranting clinical attention, the likelihood of clonality devoid of clinical significance is small. Furthermore, the lack of significant abnormalities detected amongst controls who received chemotherapy, and at least one presumed constitutional abnormality (patient 27), mitigates against such concerns.
In conclusion, this study indicates that an in-depth unbiased whole-genome analysis can prove useful if judiciously applied on a larger scale. Forty-two percent of cases with indeterminate morphologic findings were reclassified as clonal cytopenia(s) of undetermined significance based on the combined array findings. An additional 20% of known MDS with essentially normal cytogenetics (classified as ‘good’ or ‘very good’ cytogenetic risk category) had major cryptic chromosomal aberrations detected by combined array. In contrast, combined array findings in MDS cases with abnormal cytogenetics did not yield considerable new information in this series. In addition, combined array results from control cases (patients seen with an alternative malignant diagnosis) are described, and are comparable to the pattern seen in the marrow of patients with peripheral cytopenias, but no morphologic dysplasia. Implementation of combined array analysis detects clonal hematopoiesis in a substantial fraction of patients where there is clinical suspicion for MDS but routine cytogenetic analyses are normal and morphologic findings are not diagnostic.
References
Brunning RS, Orazi A, Germing U et al. Myelodysplastic syndromes/neoplasms, overview. In: Swerdlow H, Campo E, Harris NL (ed). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th edn. IARC Press: Lyon, France, 2008, pp 88–93.
Leone G, Mele L, Pulsoni A et al. The incidence of secondary leukemias. Haematologica 1999;84:937–945.
Fenaux P . Chromosome and molecular abnormalities in myelodysplastic syndromes. Int J Hematol 2001;73:429–437.
Haase D, Germing U, Schanz J et al. New insights into the prognostic impact of the karyotype in MDS and correlation with subtypes: evidence from a core dataset of 2124 patients. Blood 2007;110:4385–4395.
Orazi A, Czader MB . Myelodysplastic syndromes. Am J Clin Path 2009;132:290–305.
Pozdnyakova O, Miron PM, Tang G et al. Cytogenetic abnormalities in a series of 1029 patients with primary myelodysplastic syndromes: a report from the US with a focus on some undefined single chromosomal abnormalities. Cancer 2008;113:3331–3340.
Jiang H, Xue Y, Wang Q et al. The utility of fluorescence in situ hybridization analysis in diagnosing myelodysplastic syndromes is limited to cases with karyotype failure. Leuk Res 2012;36:448–452.
Pitchford CW, Hettinga AC, Reichard KK . Fluorescence in situ hybridization testing for −5/5q, −7/7q, +8, and del(20q) in primary myelodysplastic syndrome correlates with conventional cytogenetics in the setting of an adequate study. Am J Clin Path 2010;133:260–264.
Shaffer LG, Ballif BC, Schultz RA . The use of cytogenetic microarrays in myelodysplastic syndrome characterization. Methods Mol Biol 2013;973:69–85.
Evers C, Beier M, Poelitz A et al. Molecular definition of chromosome arm 5q deletion end points and detection of hidden aberrations in patients with myelodysplastic syndromes and isolated del(5q) using oligonucleotide array CGH. Genes Chromosomes Cancer 2007;46:1119–1128.
Kolquist KA, Schultz RA, Furrow A et al. Microarray-based comparative genomic hybridization of cancer targets reveals novel, recurrent genetic aberrations in the myelodysplastic syndromes. Cancer Genet 2011;204:603–628.
O'Keefe CL, Tiu R, Gondek LP et al. High-resolution genomic arrays facilitate detection of novel cryptic chromosomal lesions in myelodysplastic syndromes. Exp Hematol 2007;35:240–251.
Paulsson K, Heidenblad M, Strombeck B et al. High-resolution genome-wide array-based comparative genome hybridization reveals cryptic chromosome changes in AML and MDS cases with trisomy 8 as the sole cytogenetic aberration. Leukemia 2006;20:840–846.
Starczynowski DT, Vercauteren S, Telenius A et al. High-resolution whole genome tiling path array CGH analysis of CD34+ cells from patients with low-risk myelodysplastic syndromes reveals cryptic copy number alterations and predicts overall and leukemia-free survival. Blood 2008;112:3412–3424.
Thiel A, Beier M, Ingenhag D et al. Comprehensive array CGH of normal karyotype myelodysplastic syndromes reveals hidden recurrent and individual genomic copy number alterations with prognostic relevance. Leukemia 2011;25:387–399.
Volkert S, Haferlach T, Holzwarth J et al. Array CGH identifies copy number changes in 11% of 520 MDS patients with normal karyotype and uncovers prognostically relevant deletions. Leukemia 2016;30:259–261.
Cluzeau T, Moreilhon C, Mounier N et al. Total genomic alteration as measured by SNP-array-based molecular karyotyping is predictive of overall survival in a cohort of MDS or AML patients treated with azacitidine. Blood Cancer J 2013;3:e155.
Gondek LP, Dunbar AJ, Szpurka H et al. SNP array karyotyping allows for the detection of uniparental disomy and cryptic chromosomal abnormalities in MDS/MPD-U and MPD. PloS One 2007;2:e1225.
Gondek LP, Haddad AS, O'Keefe CL et al. Detection of cryptic chromosomal lesions including acquired segmental uniparental disomy in advanced and low-risk myelodysplastic syndromes. Exp Hematol 2007;35:1728–1738.
Gondek LP, Tiu R, Haddad AS et al. Single nucleotide polymorphism arrays complement metaphase cytogenetics in detection of new chromosomal lesions in MDS. Leukemia 2007;21:2058–2061.
Gondek LP, Tiu R, O'Keefe CL et al. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood 2008;111:1534–1542.
Hahm C, Mun YC, Seong CM et al. Single nucleotide polymorphism array-based karyotyping in acute myeloid leukemia or myelodysplastic syndrome with trisomy 8 as the sole chromosomal abnormality. Acta Haematol 2013;129:154–158.
Heinrichs S, Kulkarni RV, Bueso-Ramos CE et al. Accurate detection of uniparental disomy and microdeletions by SNP array analysis in myelodysplastic syndromes with normal cytogenetics. Leukemia 2009;23:1605–1613.
Mohamedali A, Gaken J, Twine NA et al. Prevalence and prognostic significance of allelic imbalance by single-nucleotide polymorphism analysis in low-risk myelodysplastic syndromes. Blood 2007;110:3365–3373.
O'Keefe C, McDevitt MA, Maciejewski JP . Copy neutral loss of heterozygosity: a novel chromosomal lesion in myeloid malignancies. Blood 2010;115:2731–2739.
Tiu RV, Gondek LP, O'Keefe CL et al. Prognostic impact of SNP array karyotyping in myelodysplastic syndromes and related myeloid malignancies. Blood 2011;117:4552–4560.
Yi JH, Huh J, Kim HJ et al. Genome-wide single-nucleotide polymorphism array-based karyotyping in myelodysplastic syndrome and chronic myelomonocytic leukemia and its impact on treatment outcomes following decitabine treatment. Ann Hematol 2013;92:459–469.
Arenillas L, Mallo M, Ramos F et al. Single nucleotide polymorphism array karyotyping: a diagnostic and prognostic tool in myelodysplastic syndromes with unsuccessful conventional cytogenetic testing. Genes Chromosomes Cancer 2013;52:1167–1177.
Stevens-Kroef MJ, Hebeda KM, Verwiel ET et al. Microarray-based genomic profiling and in situ hybridization on fibrotic bone marrow biopsies for the identification of numerical chromosomal abnormalities in myelodysplastic syndrome. Mol Cytogenet 2015;8:33.
Kwok B, Hall JM, Witte JS et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood 2015;126:2355–2361.
Steensma DP, Bejar R, Jaiswal S et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015;126:9–16.
Petersen AK, Ahmad A, Shafiq M et al. Deletion 1q43 encompassing only CHRM3 in a patient with autistic disorder. Eur J Med Genet 2013;56:118–122.
Deshpande AJ, Chen L, Fazio M et al. Leukemic transformation by the MLL-AF6 fusion oncogene requires the H3K79 methyltransferase Dot1l. Blood 2013;121:2533–2541.
Manara E, Baron E, Tregnago C et al. MLL-AF6 fusion oncogene sequesters AF6 into the nucleus to trigger RAS activation in myeloid leukemia. Blood 2014;124:263–272.
Jankowska AM, Szpurka H, Tiu RV et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood 2009;113:6403–6410.
Langemeijer SM, Kuiper RP, Berends M et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 2009;41:838–842.
Gough SM, Slape CI, Aplan PD . NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 2011;118:6247–6257.
De Keersmaecker K, Porcu M, Cox L et al. NUP214-ABL1-mediated cell proliferation in T-cell acute lymphoblastic leukemia is dependent on the LCK kinase and various interacting proteins. Haematologica 2014;99:85–93.
Laurie CC, Laurie CA, Rice K et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet 2012;44:642–650.
Jacobs KB, Yeager M, Zhou W et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet 2012;44:651–658.
Bejar R, Stevenson K, Abdel-Wahab O et al. Clinical effect of point mutations in myelodysplastic syndromes. New Engl J Med 2011;364:2496–2506.
Cazzola M, Della Porta MG, Malcovati L . The genetic basis of myelodysplasia and its clinical relevance. Blood 2013;122:4021–4034.
Haferlach T, Nagata Y, Grossmann V et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014;28:241–247.
Genovese G, Kahler AK, Handsaker RE et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. New Engl J Med 2014;371:2477–2487.
Jaiswal S, Fontanillas P, Flannick J et al. Age-related clonal hematopoiesis associated with adverse outcomes. New Engl J Med 2014;371:2488–2498.
Xie M, Lu C, Wang J et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014;20:1472–1478.
Cargo CA, Rowbotham N, Evans PA et al. Targeted sequencing identifies patients with pre-clinical MDS at high risk of disease progression. Blood 2015;126:2362–2365.
Acknowledgements
We acknowledge the staff at the microarray CGH laboratory at URMC for their technical assistance, Andrea Baran for statistical assistance, and Dr John Bennett, M.D., Department of Pathology and Laboratory Medicine for critical comments.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Evans, A., Ahmad, A., Burack, W. et al. Combined comparative genomic hybridization and single-nucleotide polymorphism array detects cryptic chromosomal lesions in both myelodysplastic syndromes and cytopenias of undetermined significance. Mod Pathol 29, 1183–1199 (2016). https://doi.org/10.1038/modpathol.2016.104
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.104
This article is cited by
-
Clinical Applications of Chromosomal Microarray Testing in Myeloid Malignancies
Current Hematologic Malignancy Reports (2020)
-
Identify latent chromosomal aberrations relevant to myelodysplastic syndromes
Scientific Reports (2017)