Abstract
High-risk human papillomavirus (HPV) types cause cervical lesions of varying severity, ranging from transient productive infections to high-grade neoplasia. Disease stratification requires the examination of lesional pathology, and possibly also the detection of biomarkers. P16INK4a and MCM are established surrogates of high-risk HPV E6/E7 activity, and can be extensively expressed in high-grade lesions. Here we have combined these two cellular biomarkers with detection of the abundant HPV-encoded E4 protein in order to identify both productive and transforming lesions. This approach has allowed us to distinguish true papillomavirus infections from similar pathologies, and has allowed us to divide the heterogeneous CIN2 category into those that are CIN1-like and express E4, and those that more closely resemble nonproductive CIN3. To achieve this, 530 lesional areas were evaluated according to standard pathology criteria and by using a multiple staining approach that allows us to superimpose biomarker patterns either singly or in combination onto an annotated hematoxylin and eosin (H&E) image. Conventional grading of neoplasia was established by review panel, and compared directly with the composite molecular pathology visualized on the same tissue section. The detection of E4 coincided with the onset of vacuolation, becoming abundant in koilocytes as the MCM marker declined and cells lost their defined nuclear margins as visualized by standard H&E staining. Of the dual marker approaches, p16INK4a and E4 appeared most promising, with E4 generally identifying areas of low-grade disease even when p16INK4a was present. Extensive p16INK4a expression usually coincided with an absence of E4 expression or its focal retention in sporadic cells within the lesion. Our results suggest that a straightforward molecular evaluation of HPV life-cycle deregulation in cervical neoplasia may help improve disease stratification, and that this can be achieved using complementary molecular biomarker pairs such as MCM/E4 or, more promisingly, p16INK4a/E4 as an adjunct to conventional pathology.
Similar content being viewed by others
Main
The International Agency for Research on Cancer states that 4.8% (610 000) of new cancers occurring in 2008 worldwide were attributable to human papillomavirus (HPV) infection, with substantially higher incidence and mortality rates in developing versus developed countries.1, 2 HPV has a causal etiology for cancers of the cervix uteri, penis, vulva, vagina, anus, and oropharynx, including the base of the tongue and tonsils.3, 4 To date, >200 different types of HPV have been identified on the basis of sequence analysis (http://pave.niaid.nih.gov/#home,5), with each type showing a defined epithelial tropism and disease association.3, 6 High-risk α-HPV (high-risk HPV) types are the primary cause of cervical neoplasia and cervical cancer, a disease that affects over half a million women per year worldwide.3, 7, 8
In squamous cervical epithelium, the mitotically active basal and parabasal cells occupy the cell layers immediately above the basal lamina.9 As cells enter the mid-zone, they begin to mature, and show a general increase in the volume of clear vacuolated cytoplasm containing glycogen. The differentiated superficial cells are characterized by larger areas of cytoplasm and smaller pyknotic nuclei,10 with the HPV life cycle being linked to the differentiation status of this epithelium. HPV infects actively dividing basal cells during wound healing,11 with the viral genome maintaining itself episomally at a low copy number. In the lower epithelial layers, high-risk-HPV express E6 and E7 proteins in order to stimulate cell proliferation and to ensure that differentiating cells are retained in the cell cycle.12, 13 These viral proteins stimulate the synthesis of cellular proteins necessary for S-phase entry, allowing the replication of viral episomes and initiation of the productive viral phase. As virus-infected cells approach the epithelial surface, an increase in late promoter activity leads to an elevation in replication (E1, E2), and accessory proteins (e.g., E4), followed by the assembly of infectious virions in the superficial cell layers.3 Current thinking suggest that the levels and/or activity of E6 and E7 are lower in CIN1 (low-grade squamous intraepithelial lesion (LSIL) or mild dysplasia), where basaloid cells occupy the lower third of epithelium, than in CIN2/3 (high-grade squamous intraepithelial lesion (HSIL) or moderate to severe dysplasia), where such cells occupy from two-thirds to the full thickness of the epithelium.14 Such deregulated viral gene expression is thought to underlie pathologic phenotype, and predisposes the cell to the accumulation of genetic errors and the eventual progression to cancer.14 High-grade CIN (CIN2/3; HG-CIN) lesions often do not support a productive virus life cycle, but represent a transforming viral phase or abortive virus life cycle, even though the majority of cells within the lesion still maintain intact viral episomes.15, 16 Most cervical HPV infections are cleared within 1–2 years. However, women with persistent high-risk HPV infections can develop HG-CIN.17, 18, 19 Current treatments for HG-CIN focus on eliminating abnormal HPV-infected precancerous cells by surgical excision. The use of molecular criteria that distinguish lesions supporting an abortive or transforming rather than a productive phase could thus be of value in clinical practice.20
Our understanding of the molecular biology of HPV infection and the organization of the HPV life cycle during cancer progression provides a rational basis for marker selection.21 A major class of biomarkers useful in cervical screening represents proteins activated as a consequence of expression of the viral oncogenes E6 and E7. E7 associates with the Rb cell cycle regulator and releases the E2F transcription factor that subsequently activates genes necessary for DNA replication.22 The most extensively evaluated cell cycle markers include p16INK4a, MCM (minichromosome maintenance protein), and Ki-67.23, 24, 25 When present in the upper layers of cervical lesions, these proteins can be regarded as surrogate markers of viral oncogene expression.26, 27 In low-grade cervical lesions, such proteins are typically confined to the lower epithelial layers, but extend into the higher epithelial layers in HSIL. Marked inter- and intra-observer variability even on histology indicates the need for biomarkers that are specific, sensitive, and reproducible.23, 24
The purpose of this study was to correlate the two most widely used molecular surrogates of E6/E7 expression and life-cycle deregulation (p16INK4a and MCM) with detection of the abundant HPV E4 protein that is highly expressed in productive HPV infections and that marks the initiation of the late stage of the virus life cycle. Previous studies have suggested that these two distinct classes of biomarker are complementary and that they could be used together to establish grade of neoplasia and to distinguish virus infections from other similar pathologies that may have a different prognosis.12, 28 In this study we have correlated the expression of all three biomarkers with the precise pathology characteristics that are currently used to establish lesion grade, in order to establish whether a combined molecular and standard pathology approach might offer advantages over conventional methods in the future. To do this we have carried out multiple biomarker stains on tissue sections containing different grades of neoplasia, followed by the digital overlay of each biomarker pattern either singly or in combination onto the hematoxylin and eosin (H&E) pathology. Our results indicate that a dual biomarker approach using E4 and p16INK4a can distinguish HPV-associated CIN1 from other, probably nonviral pathologies, and may be used to divide the CIN2 group according to the extent of life-cycle deregulation.
Materials and methods
Clinical Samples
Loop electrosurgical excision procedure (sometimes referred to as LEEP) and hysterectomy specimens from women treated at the Jagiellonian University Medical College, Krakow, Poland (103 biopsy specimens), as well as punch biopsies taken from women during colposcopy at the Gynaecological outpatient clinic of Hospital Clínic, Barcelona, Spain, and Reinier de Graaf Groep, Voorburg, the Netherlands (34 biopsy specimens), were used in this study.29 Patient data for all the samples were anonymized. All biopsies were fixed in buffered formalin and embedded in paraffin.
Selection of Cervical Biopsy Sections and Whole-Tissue PCR
A total of 25 serial sections of 5 μm were cut from each formalin-fixed, paraffin-embedded histology specimen. Sections 8 and 15 were taken for whole-tissue section PCR analysis and analyzed separately, and sections 12, 13, and 14 were mounted on Laser Capture Microscopy membrane slides (Zeiss, Cambridge, UK). Slide 11 was used for E4 Mab and MCM2 immunohistochemistry followed by p16INK4a and H&E staining. Slide 10 was used for pan E4 monoclonal and MCM2 immunohistochemistry followed by H&E staining. Extensive measures were implemented during sectioning to prevent cross-contamination.
Immunohistochemical Staining
E4 detection was carried out using both TVG 405 (a HPV16-, 18-, 31-, 35-, and 45-specific E4 Fab28) and a newly developed pan-specific E4 monoclonal antibody (FH1.1) reactive against the high-risk HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 67, and 70 (Zhonglin Wu, Heather Griffin, Yasmina Soneji and John Doorbar, manuscript in preparation), depending on the HPV types present. Although TVG 405 was used for the majority of the stains, both antibodies produce equivalent staining patterns. MCM2 and p16INK4a immunohistochemistry was performed on all cervical biopsy sections according to standard procedures. For epitope retrieval, slides were incubated in solution D pH 9.0 (Dako, Glostrup, Denmark) for 10 min at room temperature before autoclaving for 2 min at 121 °C. The HPV anti-E4 Fab TVG 405 was directly conjugated to Alexa 488 and was diluted 150-fold before use. The concentrated monoclonal E4 antibody (FH1.1) was diluted 100-fold and detected using an anti-mouse Alexa 488-conjugated antibody (dilution 1:150, Vector, Peterborough, UK). E4 staining was combined with MCM2 using a primary rabbit polyclonal antibody to MCM2 (Abcam, Cambridge, UK). MCM antibody detection was carried out using an anti-rabbit Alexa 594 conjugated antibody (dilution 1:150, Vector). Nuclear counterstain was performed with 4’-6-diamidino-2-phenylindole (DAPI, 1 mg/ml 200- to 500-fold diluted, Sigma, St Louis, MO, USA) before mounting in Citifluor medium (Agar Scientific, Essex, UK) for fluorescence scanning and digital imaging on a Pannoramic Slide Scanner (3D Histotech, Hungary).
Sections that had been stained with the Fab fragment to HPV E4 and a rabbit polyclonal antibody to MCM2 were then stained using a mouse primary antibody to p16INK4a clone JC8 diluted 1:20 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) that was detected using an anti-mouse biotinylated antibody (dilution 1:150, Vector) followed by development using an ABC kit (Vector) and red AEC-reagent (Sigma). After scanning and digital imaging of the p16INK4a stain (using the Pannoramic Slide Scanner (3D Histotech) as described above), slides were H&E stained with Carazzi’s hematoxylin ( × 2) before rescanning under bright-field illumination.
For p16INK4a, strong nuclear and cytoplasmic staining was considered as positivity. For MCM2, strong nuclear staining was considered as positivity. The p16INK4a and MCM2 results were reported in a semiquantitative manner as negative or focal staining, staining up to one-third of the epithelium, staining up to two-thirds of the epithelium, or staining above two-thirds of the epithelium. Scoring was based on the highest category within an annotated area. The immunohistochemistry staining and reading was carried out by three individuals (HG, YS, and RvB).
HPV DNA Detection and Laser Capture Microdissection
HPV typing was carried out on all cases using a PCR/Line Probe Assay (SPF10-PCR-DEIA-LiPA25, Version 1, Labo Biomedical Products BV, Rijswijk, The Netherlands) as described previously30, 31 in order to assign HPV type(s) to a lesion. This is referred to in the text as whole-tissue section PCR. In CIN1 areas that were E4 negative, an adjacent section was analyzed by laser capture microdissection PCR in order to establish whether HPV DNA was detectable in the lesional area.32 Fluorescence in situ hybridization (FISH) was carried out on a subset of these E4-negative lesions (including all the total-agreement CIN1 cases) to confirm that E4 negativity correlated with an absence of genome amplification.12, 33 Sequence analysis of the SPF10 regions was done for three of the laser capture microdissection regions that remained untypeable by PCR/Line Probe Assay (LiPA25) as described previously.34 We excluded cases with HPV types that were undetectable by the antibodies used in this study.
Pathological Diagnosis and Grading
Initial diagnosis and the grading of discrete areas with different pathologies was carried out on the H&E-stained section according to standard criteria by an expert pathologist at UCL (London, UK). Annotated regions were classified as ‘non-CIN’, when HPV-associated pathology was absent, but where other histological changes, including immature metaplasia and squamous hyperplasia, were seen. When putative HPV-associated changes were noticed, they were classified either as CIN1, CIN2, CIN3, or a combination thereof (e.g., CIN1/2, CIN2/3). Grading was then independently reassessed by two expert pathologists using the same tissue section. Discrete areas within the tissue section were examined in this study, because we wanted to correlate the precise relationship between markers of infection and associated pathology, although we appreciate that in routine practice, the highest grade of disease on the tissue section must be used to determine treatment options. If all pathologists agreed on the diagnosis of disease severity, it was considered as total agreement. If two-thirds of the pathologists agreed, it was considered consensus agreement. Furthermore, all areas were scored for the presence of koilocytosis (i.e., superficial cells with perinuclear atypia and cytoplasmic cavitation (see p209 of Kurman et al10), or the presence of vacuolation, when not all the koilocyte characteristics were apparent. The pathologists were blinded to the HPV status and immunohistochemistry results.
Results
Development of an Overlay Approach for Pathology/Biomarker Correlation
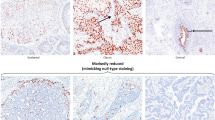
In order to correlate the expression patterns of multiple biomarkers with disease pathology, we first developed an ‘image-overlay’ approach. Using this methodology, the distribution of key molecular markers were imaged and captured separately following immunofluorescence staining or immunohistochemistry (Figure 1a). Our marker panel included p16INK4a, which is an established marker of deregulated high-risk HPV gene expression, and MCM, which identifies cells ‘in cycle’. Both are considered as surrogates of E6/E7 expression when present in HPV-associated cervical neoplasia. An important aspect of this study is the use of such markers in combination with the E4 biomarker that represents a separate category of marker that marks the onset of productive infection. The staining and image capture regime is shown in Figure 1a. At the end of this procedure, the tissue section was cleared of visible biomarker staining as part of the H&E staining regime in order to allow precise visualization of the pathology features used in conventional diagnosis. To accurately correlate molecular biomarker patterns with disease pathology, all of the H&E-stained slides were subsequently subjected to pathology review, and discrete lesional areas exhibiting distinct pathologies were annotated as shown in Figure 1a. Individual biomarkers were then digitally superimposed onto the annotated H&E image in order to establish the molecular pathology of disease. The images shown in Figure 1b shows the distribution of E4 (colored green) in regions of low-grade disease, the distribution of MCM (colored red), which marks cells in cycle, and the distribution of p16INK4a (colored brown) that is most prominent in the regions of HG-CIN. As the molecular biomarker stains and H&E are all carried out on the same tissue section, individual markers can be overlayed in different combinations to fully understand how HPV activity and cellular gene expression relates to conventional pathology. The complementarity of the HPV E4 biomarker (green) and surrogate markers of E6/E7 expression (p16INK4a (brown) and MCM (red)) is shown in the low-power images in Figure 1b, as are the distinct biomarker distributions seen in low- and high-grade disease areas. During the course of the study, 530 areas with discrete cervical pathologies (as shown in Figure 1a) were identified and analyzed using the biomarker overlay approach.
Overlay of biomarker patterns onto annotated hematoxylin and eosin pathology. (a) Individual tissue sections were subject to immunofluorescence staining to detect the biomarkers E4 using a TVG405-Alexa 488 conjugate (green), MCM using an anti-mouse Alexa 594 secondary antibody (red), and cellular DNA using 4’,6-Diamidino-2-Phenylindole (DAPI; blue). Individual stains were recorded digitally at high resolution (lower panels) before the section was processed for p16INK4a staining and development in 3_Amino_9_ethylcarbazole (AEC; brown). The AEC image was digitally captured, before the tissue section was cleared of the AEC substrate and stained with Carazzi’s ( × 2) hematoxylin and eosin. All H&E-stained images were then examined independently by three pathologists, and regions with discrete pathology phenotypes recorded. The primary categories of HPV-associated pathology comprised CIN1, CIN2, and CIN3, with the general term ‘non-CIN’ being used to encompass a variety of non-HPV-associated pathologies such as inflammation and metaplasia. A ‘normal’ classification was given when there was no histological abnormality. (b) The three-color immunofluorescence stain is shown on the left. Single channel colored images (collected as described in (a)) were extracted from the immunofluorescence image or from the p16INK4a AEC stain and were superimposed onto the H&E images (under the heading ‘Pathology overlay’). The simple dual marker molecular pathologies (i.e., not overlayed onto the H&E image) are shown on the right to reveal the relative distributions of E4/MCM and E4/p16INK4a (under the heading ‘Dual marker molecular pathology). The image shown is typical of those used to prepare the more detailed images of neoplasias used in subsequent figures.
Pathology Agreement
Of the 530 annotated regions examined in this study, all pathologists agreed on the diagnosis in 146 instances, whereas in 282 areas there was a consensus by two-thirds of the pathologists. In all, 102 lesional areas were the subject of total disagreement, with pathologists disagreeing as to the precise CIN grade (e.g., CIN1 or CIN2, or CIN2 or CIN3) or whether diagnosis should be CIN1 or non-CIN (i.e., metaplasia, inflammation, and so on). In total, 276 lesional areas (52%) were classified as CIN1 by at least one pathologist. Only in 25/276 (5%) areas was there total agreement on the CIN1 diagnosis, however, with an additional 8 areas receiving a range of diagnosis comprising CIN1/2. A total of 284 lesional areas were classified as CIN2 by at least one pathologist, but in only 12/284 (4%) areas did all three pathologists agree on the diagnosis. In six of these, one of the pathologists used either a CIN2/CIN3 (four areas) or a CIN1/CIN2 (two areas) classification. A total of 311 individual areas were classified as CIN3 by at least one pathologist, but in only 68 areas was there total agreement on the CIN3 diagnosis among all three pathologists. Of these, 23 received a diagnosis of CIN2/CIN3 by one of the pathologists. The areas with total agreement were subsequently used to establish the specific criteria that might allow the use of molecular markers as surrogates of CIN grading. HPV typing analysis revealed that the majority of cases (90%) contained only a single HPV type by whole-tissue section PCR, and encompassed single infections by HPV types 16, 18, 31, 33, 35, 39, 51, 52, 53, 56, 58, 59, and 66. No additional types were detected in cases with multiple infections that contained between two and five HPV types, all of which could be detectable using the combination of E4 antibodies outlined in this study.
Total-Agreement CIN1 Generally Has a Well-Defined Molecular Pathology Pattern
HPV-induced changes are considered pathognomonic of LSILs, and of these the most significant is nuclear atypia characterized by variation in nuclear size, hyperchromasia, and irregularity or wrinkling of the nuclear membrane. Additional characteristics of HPV infection include acanthosis and koilocytosis. The 33 CIN1 regions examined here had recognizable characteristics, with koilocytosis and clear evidence of cytoplasmic vacuolation in 32 of the areas (see Figure 2ai and ii). The majority of these ‘total-agreement’ CIN1 lesions (i.e., 27 out of 33 (82%)) showed expression of the E4 biomarker (Figure 2ai and ii), although in 3 areas there was tissue damage at the epithelial surface that occurred during the biopsy procedure or during histologic processing. In contrast to the unambiguous CIN1 shown in Figure 2ai and ii, the lesion shown in Figure 2aiii was diagnosed as CIN1/basal cell hyperplasia by one of the three pathologist reviewers. HPV52 was detected within the tissue by laser capture microdissection, but this was at the limits of detection compared with the productive CIN1 cases described above, requiring DNA sequencing to confirm the infecting type as HPV52. Only five other CIN1 lesions were found to be devoid of E4, with one of these being reclassified as higher grade (CIN3) following visualization of mitotic figures in the upper epithelial third on pathology review, as well as nuclear atypia in the basal layer despite a low-grade pathology in the mid-epithelial layers. The presence of p16INK4a and MCM2 expression in the upper two-thirds of the epithelium, and the absence of E4 supported this revised grading. As reported previously, the presence of E4 correlated closely with the onset of viral genome amplification as visualized by FISH.6, 12, 33 Viral genome amplification was not seen in any of the six E4-negative CIN1 areas described here. Two of the CIN1 E4-negative areas were tangentially sectioned and thus difficult to interpret, whereas one showed clear inflammation. Interestingly, the remaining E4-negative CIN1 was among those classified as CIN1/2 by one of the pathologists (shown in Figure 2biii) and differed markedly from the above examples in containing clearly identifiable koilocyte-like cells that are suggestive of productive HPV infection (Figure 2biii), but only limited MCM2 and p16INK4a expression that was restricted to the basal and suprabasal layers (MCM2 staining shown in Figure 2biii). In the koilocyte-like cells (arrowed in Figure 2biii), there was no evidence of cell-cycle entry, viral genome amplification (as determined by FISH; data not shown), or the presence of E4 (see Figure 2biii), distinguishing this lesion from the majority of productive CIN1 that were E4 positive (see Figure 2bi and ii), and that could be confirmed as productive HPV infections. Although HPV type 33 could be detected in this lesion by laser capture microdissection, it is worth noting that HPV can sometimes also be detected in apparently normal cervical tissue as an asymptomatic or latent infection,32, 35 and that HPV positivity in CIN1 can be between 50 and 70%.36 In the absence of a definitive pathognomic biomarker pattern or other evidence of viral gene expression, viral causality remains uncertain.
Distribution of the HPV_E4, p16INK4a and MCM2 biomarkers in lesions unambiguously classified as CIN1. (a) Biomarker patterns typically associated with CIN1. The E4/MCM (green/red) biomarker patterns are shown in the immunofluorescence image to the left of the figure. To facilitate comparison with lesional pathology, the E4/MCM (green/red) and p16INK4a (brown) biomarkers are overlayed onto (and shown alongside) the basic H&E stain in the central part of the figure. The E4/MCM (green/red) and E4/p16INK4a (green/brown) biomarker overlays are shown separately to the right of the figure so that their relative distributions can be observed. The biomarker patterns seen in HPV-associated CIN1 are described below. (ai) Most CIN1 are productive infections, with MCM (red) reaching and extending into the E4-expressing layers (green). In this lesion, MCM and p16INK4a have broadly similar distributions. (aii) Although p16INK4a (brown) and MCM (red) expression can have different distributions, MCM again extends into the E4-positive layers (green). In this lesion, MCM extends higher into the epithelial layers than p16INK4a. (aiii) A small number of consensus CIN1 (6/33) failed to show E4 (green) biomarker expression. p16INK4a (brown) expression extends into the upper epithelial layers and is more extensive than MCM (red). (b) Correlation of biomarker patterns with pathology. The E4/MCM (green/red) biomarker distribution in consensus CIN1 are shown as immunofluorescence images to the left of the figure, and overlayed onto H&E pathology toward the center. The higher magnification shown on the right allows correlation of specific pathology characteristics with the accumulation of E4 and the decline of MCM. (bi) In the majority of CIN1, the E4 (green) biomarker accumulates during the process of koilocyte formation. Arrows labeled 1 to 4 show the progressive vacuolation that accompanies E4 accumulation (beginning at arrow 2 and highest at arrow 4), and the loss of the MCM biomarker (lowest at arrow 4). (bii) Despite some differences in cell morphology and tissue architecture among CIN1, the distribution of biomarkers described above (bi) is conserved. (biii) In one (out of 33) consensus CIN1, koilocyte-like cells were present but were devoid of markers of productive infection, including the E4 (green) biomarker and associated MCM (red) staining.
HPV_E4 Accumulation in CIN1 Occurs during Vacuolation and Koilocyte Development
To correlate virus-associated pathology with biomarker presence more precisely, the E4/MCM2 biomarker pair was overlayed onto H&E-stained images in all of the low-grade lesional areas described above. This analysis revealed a reproducible relationship between marker presence and viral pathology (Figure 2b). Cells with well-defined nuclear margins, before the first appearance of E4, stained strongly with MCM2 (cells labeled ‘1’ in Figure 2bi and ii). The appearance of E4 coincided with the first signs of cellular vacuolization in HPV-associated CIN, with vacuolization becoming more apparent in these E4-positive cells upon further differentiation (cells labeled ‘2’ in Figure 2bi and ii). E4 was typically more apparent in koilocytic cells however (labeled ‘3’ in Figure 2b) that we suspect arise from these first E4-positive vacuolated cells. The koilocytes and the cells immediately above them accumulate higher levels of the E4 biomarker and lose their well-defined nuclear margins in the H&E stain, with lower and more diffuse MCM2 expression (cells labeled ‘4’ in Figure 2bi and ii). The larger nuclear size often seen in these cells is compatible with what might be expected following cell cycle arrest in G2 that is thought to be required for genome amplification and E4 accumulation.37 Interestingly, a subset of CIN1 showed a noticeable elevation, rather than decline in MCM2 expression as the E4 biomarker appeared, with little basal cell expression (data not shown), similar to what is occasionally seen in HPV16 organotypic raft cultures.14 This is reminiscent of benign productive lesions (i.e., warts) caused by low-risk papillomaviruses. When taken alongside the variation in p16INK4a staining patterns seen in CIN1, the data suggest some variation in viral gene expression in CIN1.
These distinctive biomarker correlations were not apparent in the atypical CIN1/CIN2 (see koilocyte-like cells marked by ‘K’ in Figure 2biii). Interestingly, the nonproductive infection shown in Figure 2aiii shows (upon close inspection) only the very first signs of koilocyte formation, in agreement with its distinct molecular pathology. Although MCM2 and p16INK4a staining have very similar distributions here, in productive HPV infections such as that shown in Figure 2aii, robust MCM2 expression persists into the upper epithelial layers beyond those where p16INK4a is found. Given that MCM is a marker of E6/E7-mediated cell cycle entry in such lesions, and that cell cycle entry is required for genome amplification, this pattern fits well with our understanding of the HPV life-cycle organization. In contrast, p16INK4a is considered to be a marker of deregulated E6/E7 gene expression that may account for its more limited distribution.
Diagnostic Relevance of the HPV_E4 Biomarker in p16INK4a/MCM-Positive CIN3
In 63 cases where there was total agreement on the CIN3 diagnosis (out of 68), no E4 could be detected by any of the E4 antibodies used in this study. P16INK4a and MCM expression extended from two-thirds of the epithelium up to the surface with nuclear crowding, pleomorphism, and loss of polarity clearly visible (Figure 3ai and ii). Although absence of the E4 biomarker was by far the majority pattern in the CIN3 group, two of the E4-negative cases were also p16INK4a negative, even though MCM2 levels extended to above two-thirds of the epithelium. Among the five CIN3 areas where E4 was detected, the majority (i.e., 4 out of 5) contained only single E4-positive cells, or small clusters of such cells in isolated regions of differentiation close to the epithelial surface. These isolated cells or cell groups generally showed some evidence of koilocytosis and viral cytopathic effect, similar to what is found more extensively in low-grade disease (cells marked by arrows 1 and 2 in Figure 3c, panel B), although it was notable that not all vacuolated cells were E4 positive (cells marked by arrow ‘v’ in Figure 3c, panel A). Only in one CIN3 was E4 expression extensive, and in this case the lesional area exhibited some degree of tangential sectioning with mixed or heterogeneous pathology. MCM expression extended throughout the epithelium and facilitated the visualization of nuclear atypia, nuclear crowding (as seen in Figure 3bii), pleomorphism, and loss of normal cell polarity. P16INK4a expression was typically found to extend from between two-thirds of the epithelium up to the surface and was broadly complementary to the pattern of staining seen with E4 (Figure 3a and b).
Relevance of the HPV_E4, p16INK4a and MCM biomarkers in consensus CIN3 lesions. (a) Biomarker patterns typically associated with CIN3. The E4/MCM (green/red) and p16INK4a (brown) biomarker images are formatted as outlined in Figure 2a. The biomarker patterns seen in HPV-associated CIN3 are described below. (ai) In the majority of CIN3, the E4 (green) biomarker is absent, and both the MCM (red) and p16INK4a (brown) biomarkers extend uniformly through the full thickness of the epithelium. (aii) Despite differences in lesion size and morphology, the biomarker patterns described above (ai) were broadly similar in other CIN3. (b) Expression of the E4 biomarker is an occasional occurrence in CIN3. (bi) In a small number of CIN3 (4/68), focal areas of E4 (green) were apparent. Both p16INK4a (brown) and MCM (red) extended through the full thickness of the lesion with evidence of nuclear crowding as seen in (a). Panel (a) is shown enlarged in (c). (bii) In one case (out of 68), the E4 (green) biomarker was extensive, but was confined to regions showing low-grade pathology that lacked strong p16INK4a (brown) biomarker staining. Panel (b) is shown enlarged in (c). (c) Pathology associated with E4 expression in CIN3. The three images to the left show an enlargement of Figure 3B(i) panel A. Although vacuolated cells are sometimes apparent in CIN3 (arrows marked ‘v’), they are not necessarily associated with expression of the E4 biomarker (Figure 2bi). The pathology associated with E4 expression in CIN3 is more clearly shown in the images to the right, which is an enlargement of panel B from Figure 3B(ii). This region of low-grade pathology was contained within the CIN3 area. The pattern of vacuolation and koilocyte formation is comparable to that seen in CIN1 (Figure 2b), with E4 expression beginning at the first sign of vacuolation (arrows labeled 1) and becoming prominent as the MCM biomarker is lost (arrows labeled 2).
Division of the CIN2 Group according to HPV_E4 and p16INK4a/MCM2 Biomarker Patterns
Studies of the pathological diagnosis of CIN2 have shown that it is only poorly reproducible.38 The E4 biomarker was apparent in 7/12 (58%) CIN2 areas with total agreement, using both E4 antibodies described in this study. Staining for E4 was however often patchy, being interspersed by regions where p16INK4a and MCM clustered close to the epithelial surface (see Figure 4a). This was particularly evident when the areas designated as CIN2 were large. Of the E4-positive areas, four showed prominent koilocytosis, with E4 being expressed in a subset of koilocytes upon epithelial differentiation (Figures 4a and 5a). P16INK4a levels extended up to and above two-thirds of the epithelium. The MCM marker typically extended into the upper two-thirds of the epithelium.
Expression of HPV_E4 along with p16INK4a and MCM suggests two categories within the consensus CIN2 group. (a) HPV_E4 positivity among consensus CIN2. The E4/MCM (green/red) and p16INK4a (brown) biomarker images are formatted as outlined in Figure 2a. The biomarker patterns seen in E4-positive HPV-associated CIN2 are described below. (ai) High cell density and the expression of MCM precedes the accumulation of E4 in a subset of cells showing evidence of vacuolation. P16INK4a and MCM expression extend throughout the full thickness of the epithelium. Panel (a) is enlarged in Figure 5. (aii) Despite differences in epithelial thickness, the biomarker pattern is preserved in other consensus CIN2. (aiii) As in CIN1, some CIN2 show a more extensive expression of MCM into the upper epithelial layers when compared with p16INK4a. E4 expression is limited to cells close to the epithelial surface. Panel (b) is enlarged in Figure 5 to illustrate the correlation between pathology and biomarker patterns. (b) HPV_E4 negativity among consensus CIN2. (bi) Loss of the E4 biomarker in a proportion of CIN2 can be associated with extensive expression of the p16INK4a/MCM biomarkers throughout the thickness of the epithelium, as well as an absence of obvious differentiation at the level of pathology. Panel C is enlarged in Figure 5. (bii) MCM can extend closer throughout the epithelium more robustly than p16INK4a without the expression of the E4 biomarker. The panel shown in D is enlarged in Figure 5 to show the absence of key pathology features apparent in the E4-positive CIN2.
Expression of the HPV_E4 biomarker in CIN2 is associated with discrete regions of CIN1-like pathology. (ai) Pathology associated with E4 expression in CIN2. The six images show enlargements of the regions that are boxed in Figure 4ai and iii (H&E images). The pattern of vacuolation and koilocyte formation is similar to that seen in CIN1 (Figure 2b), with E4 expression, vacuolation, and MCM decline coinciding closely (arrows 1 and 2). (bii) The six images show an enlargement of the regions that are boxed in Figure 4bi and ii. In these lesions, vacuolated cells are sometimes apparent (arrows marked ‘v’), but are not necessarily associated with expression of the E4 biomarker (Figure 4bi and ii).
Interestingly, five of the agreed CIN2 areas were E4 negative and showed only limited signs of vacuolation and an absence of koilocytosis. One area was found to have a mitotic figure higher than two-thirds of the epithelium. Two of the E4-negative areas were in the endocervical glands, (i.e., a site that we suspect does not support the ‘normal’ productive life cycle). All four E4-negative areas showed high p16INK4a and MCM levels up to or above two-thirds of the epithelium, suggesting that at the molecular level they may in fact be CIN3-like.
Reclassification of Ambiguous Pathology using the Dual Marker Approach
To clearly assess how molecular patterns relate to pathology, we examined all 146 total-agreement pathologies according to both E4 status and distribution of p16INK4a (Figure 6a). As each pathology diagnosis was the consensus from three pathologists, the total number of individual diagnostic opinions in this group was 438 (ie 3 × 146). Lesions classified as CIN1 and that express E4 (Figure 6a; ‘CIN1’ column (green) left-most graph)) are supporting the productive phase of the virus life cycle, and typically exhibit limited p16INK4a expression. Almost all E4-positive CIN1 were however positive for p16INK4a to some extent. In contrast, the majority of CIN3 were E4 negative or had very limited expression (Figure 6a; ‘CIN3’ column (red) right-most graph) and showed a much more extensive p16INK4a distribution (i.e., extending through two-thirds of the epithelium), in agreement with their status as transforming rather than productive infections. Although numbers were limited, the total-agreement E4-negative CIN2 generally showed a more extensive p16INK4a distribution than was seen in the E4-positive group. Although clinical practice suggests that CIN2 and CIN3 could be considered together as HSIL, it appears that E4-positive ‘CIN1-like’ lesions are more common in the CIN2 group than CIN3, reflecting the heterogeneity of CIN2. The E4 and p16INK4a biomarkers were absent in nearly all cases where there was total agreement that the pathology was not HPV associated and had another cause (e.g., metaplasia, inflammation, and so on). It would therefore appear that biomarker patterns reflect the underlying pathology.
Division of cervical pathologies according to the presence and distribution of the molecular markers E4 and p16INK4a. (a) Lesional areas where there was total agreement among the panel of pathologists. The columns in each graph show the individual diagnostic opinions provided by the pathologist panel after review of the H&E-stained slides, as either non-CIN, CIN1, CIN2, or CIN3. Graphs shown in (a) include only lesional areas where there was total agreement among the pathologist panel. This standard pathology grading is stratified according to whether the diagnosed areas were subsequently found to be HPV-E4 positive (green-edged columns/left-most graph) or E4 negative (red-edged columns/right-most graph), and to what extent the p16INK4a expression extended through the epithelium. Lesional areas showing full-thickness p16 staining are indicated as dark brown columns, with lower levels of staining being shown in lighter shades of brown. Lesional areas that lacked p16INK4a staining are indicated by white columns. In general, the patterns fit with our current model of life-cycle deregulation in high-grade neoplasia, with an absence of both markers in the ‘total-agreement’ non-CIN group. As described in the text, E4 expression in CIN3 (marked by an asterisk) was typically sporadic and in a small number of cells close to the epithelial surface. (b) All lesional areas including those where there was disagreement among the panel of pathologists. The columns show the individual diagnostic opinions provided by the pathologist panel on all lesional areas, irrespective of whether there was agreement or disagreement between individual pathologists. Labeling is as described in (a) above. Because the H&E-based diagnostic opinion often differed between the individual pathologists, the pattern of the p16INK4a and E4 staining typical of non-CIN, CIN1, CIN2, and CIN3 (shown in (a)) is less apparent, with some evidence of virus infection in the ‘proposed’ non-CIN group, and an absence of biomarker staining in some ‘proposed’ CIN1.
As part of our aim was to establish how molecular biomarkers might eventually improve diagnostic accuracy, we next went on to prepare a second chart, but this time included the 384 cases where there was partial or total pathology disagreement among the panel of pathologists (Figure 6b). In this case, the total number of individual pathologies examined was 530 (384+146) and the total number of individual opinions plotted in Figure 6b was 1590 (3 × 530). In areas considered to be low grade by at least one pathologist, E4 expression was generally accompanied by low levels of p16INK4a, similar to the total-agreement group. Interestingly, a significant fraction of the E4-negative areas were found to be p16INK4a negative, and may not have a causal association with HPV. Nonviral mimics of CIN1 could include metaplasia or inflammation. The remaining E4-negative ‘CIN1’ generally showed more extensive p16INK4a than the E4-positive group.
The majority of areas that received a CIN2 classification were E4 negative, with levels of p16INK4a expression higher in this group than in the E4-positive group. E4-positive ‘CIN2’ areas were equally divided according to whether E4 expression was extensive or restricted to focal regions similar to those seen in the total-agreement CIN3 category (Figure 6a). The E4-negative CIN2 that express high levels of p16INK4a are a potentially important group that may warrant follow-up. The E4-positive CIN3 typically expressed only very limited/focal levels of E4, and generally showed extensive p16INK4a expression supporting the high-grade diagnosis.
A fraction of the areas classified as CIN2 or CIN3 by at least one pathologist lacked expression of both the p16INK4a and E4 biomarkers, and it may be that these pathologies are not HPV associated. These areas only became evident when the consensus and total disagreements were combined, and hence they can be considered as cases that are difficult to judge. Further analysis of these biomarker negatives where two of the pathologists agreed identified two CIN2 areas judged to be metaplasia or CIN3 by the third pathologist, as well as two CIN3 areas considered to be metaplasia or CIN2.
Among the areas where there was total disagreement between the panel of pathologists, 43 areas were p16INK4a negative, with two of these being E4 positive, although the extent of E4 expression was quite restricted. The majority (41) were both E4 and p16INK4a negative. One of our pathologists graded 32 of these as non-CIN, in agreement with the E4/p16INK4a patterns, whereas another pathologist identified 24 CIN1 among this group of 43 areas. In 15 out of these 24 pathology areas, a different grading (CIN1, CIN3, and non-CIN (metaplasia/inflammation)) was given by each of the three pathologists. In another seven E4/p16INK4a-negative areas, all three pathologists reported the presence of CIN, but each gave a different diagnosis (CIN1, CIN2, CIN3). Such inter-observer variability highlights the problem of reliably discriminating true HPV-associated changes from other similar pathologies such as metaplasia or inflammation, and supports our hypothesis that the staining of molecular markers in addition to conventional pathology should help to improve overall diagnostic accuracy. More specifically, our data suggest that the molecular patterns seen in the unequivocal cases (Figure 6a), and which are likely to be true indicators of CIN pathology, could be used to refine the grading of disease in case where pathology diagnosis was equivocal (Figure 6b).
Discussion
The viral etiology of cervical neoplasia has indicated an important role for HPV testing during cervical screening.1, 39 HPV negativity means that screening intervals can be extended and that there is a low risk of cancer progression,40, 41, 42, 43 whereas the detection of HPV DNA highlights a need for triage to establish the presence of active disease and to confirm its severity. The situation is complicated however by the fact that transient HPV infections that are not likely to progress to cancer are common in young women, and that the presence of HPV in a cervical cytology or whole-tissue biopsy sample does not necessarily mean the presence of a precancerous lesion, and may in some instances represent only sexual deposition and not even infection.25 As a result, there is interest in the use of biomarker approaches as an adjunct to pathology in order to confirm infection and facilitate disease stratification. Of these, p16INK4a has been most well characterized, and its presence in cervical neoplasia is generally taken as indicating deregulated HPV ‘oncogene expression’.25, 27, 44 Although the discrimination between high- and low-grade cervical disease eventually determines treatment options, robust markers of low-grade cervical pathology that can be used alongside p16INK4a are not well developed. Therefore, in this study we considered the contribution of the abundant HPV-encoded E4 protein in discriminating between high- and low-grade disease and in identifying true viral infections from regions with similar pathology. An additional class of HPV E6/E7-induced cellular protein (MCM), similar to Ki-67, was also used.45, 46, 47 We conclude from this that the detection of E4 with p16INK4a provides additional molecular detail regarding the extent of HPV gene and life-cycle deregulation, and that such a dual marker approach can help confirm viral etiology and disease status in both high- and low-grade disease.
To achieve the above goals, we used a research/evaluation methodology that allowed us to superimpose the distribution of each marker onto a standard H&E-stained pathology image, allowing us to correlate the features that are currently used for pathology diagnosis with the presence of molecular markers. During this process, particular attention was paid to individual discrete pathologies found in each lesion, in order to establish a reference of how each biomarker relates to the underlying pathology of the tissue. From these focused observations, a common pattern of p16INK4a loss and E4 appearance was apparent as the HPV-infected cell underwent the process of epithelial differentiation. In general, this correlated with the cytopathic effects that mark the onset of productive infection during the process of koilocytosis and initiation of cell vacuolization.8 Interestingly, the appearance of koilocytes was frequently characterized by the presence of abundant E4 in the upper epithelial layers, and a restriction of p16INK4a to the lower epithelial layers within a lesion. This pattern of gene expression was more reliably followed in low-grade than high-grade disease. In general, difficulties in distinguishing normal squamous epithelium from low-grade squamous intraepithelial lesions (as reported by McCluggage et al48) reflect the problem of discriminating between superficial vacuolated cells and true koilocytes. The observation that the onset of E4 expression coincides with the appearance of vacuolated cells that subsequently go on to form koilocytes as they differentiate may thus have diagnostic significance. In addition to this correlation between E4 expression and pathology, we noticed a clear and consistent difference in the distribution of MCM, which is considered to be a surrogate marker of E6/E7 expression, and p16INK4a, which is thought to be an indicator of E6/E7 deregulation (Figure 7).25, 44, 49 Again, this was more obvious in low-grade pathologies than in CIN3, where the distribution of these two E6/E7 surrogates was in most cases very similar.
Molecular principles underlying the use of p16, MCM, and E4 as HPV-associated disease biomarkers. (a) In uninfected epithelium, the cellular MCM protein (red) is usually detectable at low levels only in the basal and parabasal cell layers as a result of cell cycle stimulation by growth factors. This facilitates the phosphorylation of pRb by cyclin-dependent kinases, the release of the E2F transcription factor, and the regulated expression of MCM. During normal metaplasia or wound healing, MCM may also be detected in the upper epithelial layers. The cellular p16INK4a protein is also stimulated by E2F, but does not usually accumulate to detectable levels in uninfected epithelium. It provides feedback regulation on the activity of cyclin-dependent kinases. p16INK4a is sometimes visualized as a weak cytoplasmic stain in cells undergoing senescence (pale brown). The HPV-encoded E4 protein is never expressed in uninfected epithelium and E4 antibodies show no reactivity with cellular proteins. (bi) In HPV-infected epithelial tissue, the high-risk E6 and E7 genes (red) are expressed together from the viral early promoter (PE), and function to drive cell-cycle entry in order to allow cell proliferation and genome amplification. The high-risk E4 gene (green) is expressed from a spliced mRNA, and becomes abundant following the activation of the viral late promoter (PL) as the infected cell exits the cell cycle and commits to true differentiation. (ii) E6 and E7 are expressed at low level in the cell, but the consequences of their presence can be visualized by alterations in the presence of p16INK4a and MCM. The association of E7 with pRb leads to E2F release irrespective of growth factor stimulation. This allows MCM and also p16INK4a to accumulate to higher levels than are typically seen in uninfected epithelium where expression is dependent on cyclin-dependent kinase activation. The E7 protein also acts to increase the transcription of p16INK4a as a result of epigenetic modification of the p16INK4a promoter. In this context, p16INK4a and MCM can be used with caution as surrogate markers of E6/E7 deregulation. The viral E4 protein becomes abundant in the upper layers of HPV-infected epithelium as a result of viral late promoter activation and the cleavage of the full-length E4 protein by calpain. Calpain-cleavage exposes a C-terminal multimerization motif in E4 that allows its assembly into amyloid-like fibers. The high-level accumulation of E4 amyloid is thought to coincide with progression of the infected cell through the G2 phase of the cell cycle and eventually to cell cycle exit, explaining its appearance as MCM levels decline.
The underlying basis for the molecular biomarker patterns observed above can to a large extent be drawn from our understanding of HPV-driven neoplastic progression and normal HPV life-cycle deregulation. Thus, high levels of E6 and E7 can stimulate the appearance of p16INK4a throughout the epithelium by interfering with the normal functions of the retinoblastoma protein (pRb) and the KDM6 histone demethylase.16, 26 Interestingly, recent studies have suggested that changes in viral gene expression may underlie such phenotypic variation in CIN1 and CIN2,14 but that host genetic changes may be additionally required for the development of CIN3.15 The MCM marker, as shown previously for Ki-67, has a distinct pattern from p16INK4a in low-grade disease, presumably because its induction marks the cell cycle entry that is ordinarily stimulated by HPV to allow genome amplification (see Figure 7).12, 24, 49 In this case, cell cycle entry associated with productive infection can be distinguished from that seen in proliferating cells by the detection of E4 and MCM together. When used singly, however, the presence of markers such as MCM and Ki-67 can sometimes be difficult to interpret, as these proteins may also be upregulated during inflammation and metaplasia. The E4 biomarker has comparable limitations when used alone, but facilitates the visualization of most, if not all, HPV-associated disease areas when used in combination. The molecular regulation that leads to E4 accumulation has previously been investigated, and involves deposition of E4 in the infected cell as amyloid fibers at around the time that genome amplification begins during productive infection.50, 51, 52 Precisely why productive infection and E4 expression are retarded in high-grade disease is only poorly understood however, although our recent work suggests a link to cell cycle duration, and the accumulation of E4 only as the G2 phase of cell cycle lengthens.37, 53, 54 These preliminary results offer some initial insight into why E4 and p16INK4a produce largely complementary patterns of staining as neoplastic severity increases.
Although the pathology overlay analysis clearly shows a relationship between E4 abundance and regions of low-grade pathology, the main purposes of using such markers is to improve diagnostic accuracy and to determine the most appropriate regimen for the treatment of disease. For cervical neoplasia, treatment generally follows the diagnosis of CIN2 or higher, and typically involves surgical removal of the infected tissue and disease margins (e.g., by cone biopsy). Although this treatment is generally effective in preventing neoplastic progression, such intervention has been linked to an increased risk of preterm delivery at childbirth.55, 56 Current thinking also suggests that there is considerable heterogeneity among CIN2, and that there is a significant problem of overtreatment within this group, partly accentuated by the difficulty of reliable diagnosis.57 From the work described here, it appears that lesions designated as CIN2 by one or more pathologists can be divided into two groups on the basis of whether significant E4 expression is retained (i.e., they are CIN1-like), or whether E4 expression is lost or is restricted to focal areas close to the epithelial surface (i.e., they are CIN3-like). In general, the absence or focal expression of the E4 biomarker correlated with established pathology features of high-grade disease, including high cell density that extended toward the epithelial surface, the presence of cells with a high nuclear to cytoplasmic ratio, and a more extensive p16INK4a expression throughout the epithelium. When our molecular understanding of the HPV life cycle is considered alongside these pathology correlations, it would seem that there is a persuasive rationale for the use of a dual molecular marker approach alongside conventional pathology to establish disease severity and the associated risk of cancer.
For routine diagnostics, an additional advantage of the molecular biomarkers described here lies in their ease of use. Both p16INK4a and MCM are already available commercially for diagnostic purposes.49, 58 Interestingly, several studies have advocated the use of HPV DNA in situ hybridization alongside p16INK4a to confirm diagnosis,48, 59 a rationale that, given the correlation between E4 expression and viral genome amplification, has some similarities to the E4/p16INK4a dual biomarker approach described here. Although E4 detection reagents remain to be commercially developed, it is the most abundantly expressed HPV protein during the viral life cycle, and can be detected in lesions without the need for signal amplification systems.28, 33, 53 The recent development of a new pan-specific HPV E4 antibody that is broadly crossreactive against high-risk E4 proteins should facilitate their further evaluation. It is generally realized that distinguishing high-grade HPV-induced disease from other events such as immature metaplasia, atypical immature metaplasia (AIM), reactive/reparative changes, or atrophy from precancer is sometimes impossible on the basis of pathology alone. The use of p16INK4a immunohistochemistry and detection of high-risk HPV can assist in determining viral etiology and has shown that a proportion of these lesions are morphologically difficult CIN3.10 The situation with low-grade cervical disease is also often very difficult. In the ASCUS-LSIL Triage Study (ALTS) for instance,60 only 43% of biopsies initially classified as low-grade HPV-associated lesions were classified as low-grade disease on review, with most discrepancies being explained by an inability of the pathologist panel to reliably discriminate between HPV-associated disease, reactive squamous proliferations, and other situations where a HPV-like low-grade phenotype may be apparent (e.g., candidiasis, trichomoniasis). The vacuolated cells shown in Figure 2biii that lack markers of productive HPV infection may in fact be pseudokoilocytes. Such issues contribute to the generally low interobserver agreement in the detection of low-grade HPV-associated disease, contrasting sharply with the excellent agreement for invasive cervical lesions and even high-grade disease that are excellent and moderately good, respectively. As shown here, E4 expression is typically associated with discrete pathology features of low-grade HPV-associated disease, and it has been suggested that careful attention to cytological and histological changes can be used to discriminate between viral and nonviral pathologies.10 In reality however, such relatively subtle changes are not easy to establish under routine screening conditions, and as has been shown for all of the markers described here, their use not only marks, but also draws attention to the region of abnormality. In our study, this was apparent in one region of CIN3 that was missed by all three pathologists, but was identified as being both p16INK4a and MCM2 positive throughout the epithelium following biomarker analysis with confirmation of CIN3 status upon pathology review. In this instance, the CIN3 region comprised a small glandular lesion within a large cone biopsy.
When taken together, it appears that conventional pathology combined with the use of the E4 and p16INK4a biomarkers has advantages over pathology alone, or the use of pathology in combination with p16INK4a staining for the stratification of HPV-associated cervical neoplasia that already provides information regarding clinical outcome.61 In particular, the use of E4 facilitates the identification of low-grade viral disease where the protein is typically abundant, and distinguishes such cases from nonviral pathologies that may need different management strategies. Our data also suggest that the various and distinct expression patterns of both p16INK4a and E4 in CIN2 may allow categorization of this heterogeneous group into a CIN1-like productive infection or a CIN3-like transforming infection group according to the extent of HPV deregulation and life-cycle completion. We cannot be sure at present whether this would help in determining patient management strategies, but suspect from our data that this might be the case. The abundance of E4 and its ease of detection should facilitate a potential role during routine diagnosis and triage following cervical screening.
References
Bosch FX, Broker TR, Forman D et al. Comprehensive control of human papillomavirus infections and related diseases. Vaccine 2013;31:I1–31.
Ferlay J, Shin HR, Bray F et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893–2917.
Doorbar J, Quint W, Banks L et al. The biology and life-cycle of human papillomaviruses. Vaccine 2012;30:F55–F70.
Westra WH . The morphologic profile of HPV-related head and neck squamous carcinoma: implications for diagnosis, prognosis, and clinical management. Head Neck Pathol 2012;6:S48–S54.
de Villiers EM, Fauquet C, Broker TR et al. Classification of papillomaviruses. Virology 2004;324:17–27.
Peh WL, Middleton K, Christensen N et al. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J Virol 2002;76:10401–10416.
Cubie HA . Diseases associated with human papillomavirus infection. Virology 2013;445:21–34.
Jenkins D . Histopathology and cytopathology of cervical cancer. Dis Markers 2007;23:199–212.
Arwert EN, Hoste E, Watt FM . Epithelial stem cells, wound healing and cancer. Nat Rev Cancer 2012;12:170–180.
Kurman RJ, Ellenson LH, Ronnett BM . Blaustein's Pathology of the Female Genital Tract. Springer: New York, USA, 2010.
Pyeon D, Pearce SM, Lank SM et al. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog 2009;5:e1000318.
Middleton K, Peh W, Southern S et al. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J Virol 2003;77:10186–10201.
Bodily J, Laimins LA . Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol 2011;19:33–39.
Isaacson Wechsler E, Wang Q, Roberts I et al. Reconstruction of human papillomavirus type 16-mediated early-stage neoplasia implicates E6/E7 deregulation and the loss of contact inhibition in neoplastic progression. J Virol 2012;86:6358–6364.
Hafner N, Driesch C, Gajda M et al. Integration of the HPV16 genome does not invariably result in high levels of viral oncogene transcripts. Oncogene 2008;27:1610–1617.
Khleif SN, DeGregori J, Yee CL et al. Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc Natl Acad Sci U S A 1996;93:4350–4354.
Chen HC, Schiffman M, Lin CY et al. Persistence of type-specific human papillomavirus infection and increased long-term risk of cervical cancer. J Natl Cancer Inst 2011;103:1387–1396.
Geraets DT, van Doorn LJ, Kleter B et al. Long-term follow-up of HPV16-positive women: persistence of the same genetic variant and low prevalence of variant co-infections. PLoS One 2013;8:e80382.
Quint KD, de Koning MN, Quint WG et al. Progression of cervical low grade squamous intraepithelial lesions: in search of prognostic biomarkers. Eur J Obstet Gynecol Reprod Biol 2013;170:501–506.
Cantor SB, Atkinson EN, Cardenas-Turanzas M et al. Natural history of cervical intraepithelial neoplasia: a meta-analysis. Acta Cytol 2005;49:405–415.
Doorbar J, Cubie H . Molecular basis for advances in cervical screening. Mol Diagn 2005;9:129–142.
Roman A, Munger K . The papillomavirus E7 proteins. Virology 2013;445:138–168.
Tornesello ML, Buonaguro L, Giorgi-Rossi P et al. Viral and cellular biomarkers in the diagnosis of cervical intraepithelial neoplasia and cancer. BioMed Res Int 2013;2013:519619.
von Knebel Doeberitz M, Reuschenbach M, Schmidt D et al. Biomarkers for cervical cancer screening: the role of p16(INK4a) to highlight transforming HPV infections. Expert Rev Proteomics 2012;9:149–163.
Ikenberg H, Bergeron C, Schmidt D et al. Screening for cervical cancer precursors with p16/Ki-67 dual-stained cytology: results of the PALMS study. J Natl Cancer Inst 2013;105:1550–1557.
McLaughlin-Drubin ME, Crum CP, Munger K . Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA 2011;108:2130–2135.
Doorbar J . Papillomavirus life cycle organization and biomarker selection. Dis Markers 2007;23:297–313.
Griffin H, Wu Z, Marnane R et al. E4 antibodies facilitate detection and type-assignment of active HPV infection in cervical disease. PLoS One 2012;7:e49974.
van der Marel J, van Baars R, Quint W et al. The impact of human papillomavirus genotype on colposcopic appearance: a cross-sectional analysis. BJOG 2014;121:1117–1126.
Kleter B, van Doorn LJ, ter Schegget J et al. Novel short-fragment PCR assay for highly sensitive broad-spectrum detection of anogenital human papillomaviruses. Am J Pathol 1998;153:1731–1739.
Kleter B, van Doorn LJ, Schrauwen L et al. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol 1999;37:2508–2517.
Quint W, Jenkins D, Molijn A et al. One virus, one lesion—individual components of CIN lesions contain a specific HPV type. J Pathol 2012;227:62–71.
Doorbar J, Foo C, Coleman N et al. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 1997;238:40–52.
de Sanjose S, Quint WG, Alemany L et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol 2010;11:1048–1056.
Kalantari M, Garcia-Carranca A, Morales-Vazquez CD et al. Laser capture microdissection of cervical human papillomavirus infections: copy number of the virus in cancerous and normal tissue and heterogeneous DNA methylation. Virology 2009;390:261–267.
Guan P, Howell-Jones R, Li N et al. Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer. Int J Cancer 2012;131:2349–2359.
Davy CE, Jackson DJ, Raj K et al. Human papillomavirus type 16 E1—E4-induced G2 arrest is associated with cytoplasmic retention of active Cdk1/Cyclin B1 complexes. J Virol 2005;79:3998–4011.
Carreon JD, Sherman ME, Guillen D et al. CIN2 is a much less reproducible and less valid diagnosis than CIN3: results from a histological review of population-based cervical samples. Int J Gynecol Pathol 2007;26:441–446.
Cuzick J, Bergeron C, von Knebel Doeberitz M et al. New technologies and procedures for cervical cancer screening. Vaccine 2012;30:F107–F116.
Bulkmans NW, Rozendaal L, Voorhorst FJ et al. Long-term protective effect of high-risk human papillomavirus testing in population-based cervical screening. Br J Cancer 2005;92:1800–1802.
Elfstrom KM, Smelov V, Johansson AL et al. Long-term HPV type-specific risks for ASCUS and LSIL: a 14-year follow-up of a randomized primary HPV screening trial. Int J Cancer 2014;136:350–359.
Ronco G, Dillner J, Elfstrom KM et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet 2014;383:524–532.
Kocken M, Uijterwaal MH, de Vries AL et al. High-risk human papillomavirus testing versus cytology in predicting post-treatment disease in women treated for high-grade cervical disease: a systematic review and meta-analysis. Gynecol Oncol 2012;125:500–507.
Litjens RJ, Hopman AH, van de Vijver KK et al. Molecular biomarkers in cervical cancer diagnosis: a critical appraisal. Expert Opin Med Diagn 2013;7:365–377.
Maiorano D, Lutzmann M, Mechali M . MCM proteins and DNA replication. Curr Opin Cell Biol 2006;18:130–136.
Murphy N, Ring M, Heffron CC et al. Quantitation of CDC6 and MCM5 mRNA in cervical intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix. Mod Pathol 2005;18:844–849.
Murphy N, Ring M, Heffron CC et al. p16INK4A, CDC6, and MCM5: predictive biomarkers in cervical preinvasive neoplasia and cervical cancer. J Clin Pathol 2005;58:525–534.
McCluggage WG, Walsh MY, Thornton CM et al. Inter- and intra-observer variation in the histopathological reporting of cervical squamous intraepithelial lesions using a modified Bethesda grading system. Br J Obstet Gynaecol 1998;105:206–210.
Piaton E, Advenier AS, Carre C et al. p16(INK4a) /Ki-67 dual labelling as a marker for the presence of high-grade cancer cells or disease progression in urinary cytopathology. Cytopathology 2013;24:327–334.
McIntosh PB, Martin SR, Jackson DJ et al. Structural analysis reveals an amyloid form of the human papillomavirus type 16 E1— E4 protein and provides a molecular basis for its accumulation. J Virol 2008;82:8196–8203.
Khan J, Davy CE, McIntosh PB et al. Role of calpain in the formation of human papillomavirus type 16 E1-E4 amyloid fibers and reorganization of the keratin network. J Virol 2011;85:9984–9997.
Wang Q, Kennedy A, Das P et al. Phosphorylation of the human papillomavirus type 16 E1-E4 protein at T57 by ERK triggers a structural change that enhances keratin binding and protein stability. J Virol 2009;83:3668–3683.
Doorbar J . The E4 protein; structure, function and patterns of expression. Virology 2013;445:80–98.
Davy C, Doorbar J . G2/M cell cycle arrest in the life cycle of viruses. Virology 2007;368:219–226.
Ortoft G, Henriksen T, Hansen E et al. After conisation of the cervix, the perinatal mortality as a result of preterm delivery increases in subsequent pregnancy. BJOG 2011;117:258–267.
Armarnik S, Sheiner E, Piura B et al. Obstetric outcome following cervical conization. Arch Gynecol Obstet 2011;283:765–769.
Kinney W, Hunt WC, Dinkelspiel H et al. Cervical excisional treatment of young women: a population-based study. Gynecol Oncol 2014;132:628–635.
Henderson D, Hall L, Prpic N et al. The selection and characterization of antibodies to minichromosome maintenance proteins that highlight cervical dysplasia. J Immunol Methods 2011;370:1–13.
Galgano MT, Castle PE, Atkins KA et al. Using biomarkers as objective standards in the diagnosis of cervical biopsies. Am J Surg Pathol 2010;34:1077–1087.
Stoler MH, Schiffman M . Atypical Squamous Cells of Undetermined Significance-Low-Grade Squamous Intraepithelial Lesion Triage Study Group. Interobserver reproducibility of cervical cytologic and histologic interpretations: realistic estimates from the ASCUS-LSIL Triage Study. JAMA 2001;285:1500–1505.
Vrdoljak-Mozetic D, Krasevic M, Versa Ostojic D et al. HPV16 genotype, p16/Ki-67 dual staining and koilocytic morphology as potential predictors of the clinical outcome for cervical low-grade squamous intraepithelial lesions. Cytopathology 2013;26:10–18.
Acknowledgements
JD, HG, YS, and ZW were funded by the UK Medical Research Council. RvB was funded by the Stichting Pathologie Ontwikkeling en Onderzoek (SPOO) Foundation, The Netherlands.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Griffin, H., Soneji, Y., Van Baars, R. et al. Stratification of HPV-induced cervical pathology using the virally encoded molecular marker E4 in combination with p16 or MCM. Mod Pathol 28, 977–993 (2015). https://doi.org/10.1038/modpathol.2015.52
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2015.52
This article is cited by
-
Blimp-1 is a prognostic indicator for progression of cervical intraepithelial neoplasia grade 2
Journal of Cancer Research and Clinical Oncology (2022)
-
Use of Cap Analysis Gene Expression to detect human papillomavirus promoter activity patterns at different disease stages
Scientific Reports (2020)
-
Human papillomavirus type 16 causes a defined subset of conjunctival in situ squamous cell carcinomas
Modern Pathology (2020)
-
Characterization of cervical biopsies of women with HIV and HPV co-infection using p16ink4a, ki-67 and HPV E4 immunohistochemistry and DNA methylation
Modern Pathology (2020)
-
Paving the way for more precise diagnosis of EcPV2-associated equine penile lesions
BMC Veterinary Research (2019)