Abstract
The diagnosis and management of uterine smooth muscle tumors with uncertain malignant potential (STUMP) is often challenging, and genomic data on these lesions as well as on uterine smooth muscle lesions are limited. We tested the hypothesis that genomic profile determination by array-CGH could split STUMP into a benign group with scarce chromosomal alterations akin to leiomyoma and a malignant group with high chromosomal instability akin to leiomyosarcoma. Array-CGH genomic profile analysis was conducted for a series of 29 cases of uterine STUMP. A group of ten uterine leiomyomas and ten uterine leiomyosarcomas served as controls. The mean age was 50 years (range, 24–85) and the follow-up ranged from 12 to 156 months (average 70 months). Since STUMP is a heterogenous group of tumors with genomic profiles that can harbor few to many chromosomal alterations, we compared genomic indices in leiomyomas and leiomyosarcomas and set a genomic index=10 threshold. Tumors with a genomic index <10 were classified as nonrecurring STUMPs and those with a genomic index >10 represented STUMPs with recurrences and unfavorable outcomes. Hence, the genomic index threshold splits the STUMP category into two groups of tumors with different outcomes: a group comparable to leiomyomas and another similar to leiomyosarcomas, but more indolent. In our STUMP series, genomic analysis by array-CGH is an innovative diagnostic tool for problematic smooth muscle uterine lesions, complementary to the morphological evaluation approach. We provide an improved classification method for distinguishing truly malignant tumors from benign lesions within the category of STUMP, especially those with equivocal morphological features.
Similar content being viewed by others
Main
Smooth muscle uterine tumors include benign tumors—termed leiomyoma—that represent the most common tumor in women, and malignant tumors—such as leiomyosarcoma—that represent 1–2% of all uterine cancers and 40% of uterine sarcomas.1 These two entities are differentiated based on the ‘three features’ (cytological atypia, mitotic count, and tumor cell necrosis) morphologic approach, first proposed by investigators from Stanford in 1994.2 Although these guidelines allow to successfully classify the most conventional leiomyomas and leiomyosarcomas, some problematic lesions do not completely fulfill the criteria for malignant tumors (eg, diffuse moderate atypia but <10 mitotic figures/10 HPF, or equivocal necrosis with <10 mitotic figures/10 HPF and without atypia, or a borderline mitotic count 8–10 mitotic figures/10 HPF associated with diffuse atypia without necrosis) and are difficult to classify. A uterine smooth muscle tumor that cannot be unequivocally diagnosed as benign or malignant should be termed smooth muscle tumor with uncertain malignant potential (smooth muscle tumors with uncertain malignant potential (STUMP)), and only the outcome will confirm its benign or malignant nature.3
To complement the morphological analysis in difficult cases, a variety of ancillary laboratory techniques (p53, p16, bcl2, and ki67) have been implemented but without any significant improvement in the diagnosis.4, 5, 6, 7, 8, 9
By trying to avoid the diagnosis of STUMP, the pathologist could risk under- or over diagnosing a difficult lesion with the inherent risk of under or overtreatment, which could have dramatic consequences for the patient, especially in her reproductive age (hysterectomy precluding fertility).
To our knowledge, only a few reports have been published on STUMP,10, 11, 12 and similarly, studies on the genomic profile of leiomyomas, leiomyosarcomas are relatively scarce and cover a very-limited number of cases.13, 14, 15 Recently, a signature associated with instability and chromosomal complexity predictive of metastatic relapse in soft tissue sarcoma was published by our team.16 We demonstrated that this signature, based on expression profiles (CINSARC) was closely linked to the level of rearrangement of the tumoral genome in gastrointestinal stromal tumor, and created a way to identify a genomic signature combining the number and the type of tumoral genomic alterations evaluated by array-CGH. The genomic signature (genomic index) is a powerful predictor of metastatic relapse, as is the CINSARC signature.17 Further, this array-CGH technique is adapted to formalin-fixed paraffin-embedded tumor material and its efficacy has been demonstrated on tumor samples that have been archived for over 15 years.
Based on these findings we set up to test on a series of problematic uterine smooth muscle lesions the hypothesis that genomic profile analysis by array-CGH could split the group of borderline cases (STUMP) into two main groups consisting of1 benign tumors (leiomyomas) with a low number of chromosomal alterations and2 malignant ones (leiomyosarcomas) with a high chromosomal instability.
Materials and methods
Tumor Samples
Formalin-fixed paraffin-embedded tumor tissues from 29 uterine smooth muscle lesions diagnosed as STUMP were obtained from the pathology archives of 12 centers in France, Belgium, and Czech Republic (Institut Bergonié of Bordeaux, Université libre de Bruxelles, Katholieke Universiteit of Leuven, University of Prague, Hôpital La Croix Rousse of Lyon, Centre Jean Perrin of Clermont Ferrand, Centre JF Leclerc of Dijon, Centre A Vautrin of Nancy, Centre Paul Papin of Angers, Centre Oscar Lambret of Lille, Institut Gustave Roussy of Villejuif, and University Hospital of Montpellier) from 1999 to 2013. All cases were centrally reviewed by two gyneco-pathologists (SC and GMG). The presence of atypia, the mitotic count and the presence of tumor cell necrosis were assessed according to the Stanford criteria.2
The samples issued by the tumor archives of each department participating in the study were centralized in the Biological Resources Center of Institut Bergonié, which received the agreement from the French authorities to deliver samples for scientific research (AC-2008-812).
The control group used as a reference for the assessment of genomic index consisted of ten formalin-fixed paraffin-embedded conventional leiomyomas and 10 conventional leiomyosarcomas with unquestionable features of benignity (for leiomyomas) and malignity (for leiomyosarcomas) that were collected from three of the participating institutions (Bruxelles, Bordeaux and Dijon).
Array-Comparative Genomic Hybridization Analysis
DNA was hybridized to 8 × 60 K whole-genome Agilent arrays (G4450A; Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s protocol. The ADM-2 algorithm of the CGH Analytics v4.0.76 software (Agilent Technologies) was used to identify DNA copy number anomalies at the probe level. A low-level copy number gain was defined as a log 2 ratio >0.25 and a copy number loss was defined as a log 2 ratio <0.25. A high-level gain or amplification was defined as a log 2 ratio>1.5 and a homozygous deletion was suspected when the ratio was <−1.
The genomic index was calculated for each profile as follows: genomic index=A2/C, where A is the total number of alterations (segmental gains and losses) and C is the number of involved chromosomes.
Statistical Analysis
Metastasis-free survival was calculated by the Kaplan–Meier method from the date of initial diagnosis to the date of first metastasis or last follow-up. Overall survival using the Kaplan–Meier method was calculated from the date of diagnosis to death or last follow-up. Survival curves were compared with the log-rank test. All survival analyses were performed by using the R software version 2.14.1 and the ‘survival’ package (Terry Therneau 2-02). A Package for Survival Analysis in S. R package version 2.36–14. Multivariate analysis was done by using Cox regression with Firth’s correction (R by Meinhard Ploner and Fortran by Georg Heinze 2011). coxphf: Cox regression with Firth's penalized likelihood. R package version 1.08. http://CRAN.R-project.org/package=coxphf ).
Results
Clinico-Pathological and Genomic Data
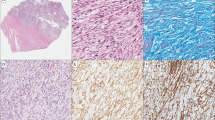
Clinico-pathological data of the 29 samples are summarized in Table 1 for the STUMP series. Morphologically, all cases were uterine smooth muscle lesions of conventional (spindle cell) type, without any epithelioid or myxoid component (Figure 1). Regarding the diagnosis of STUMP, full agreement was obtained in 7/29 cases and 22/29 had discordant diagnoses. The cause of disagreement was the appreciation of the nature of the necrosis (tumoral versus ischemic necrosis) in 16/22 cases, and the extension (focal or diffuse) and the degree of the atypia (mild, moderate, or high) in the remaining 6/22 cases. Among the latter, 1/6 tumor (case 2) presented marked nuclear atypia consisting of multinucleated giant cells in a patchy distribution with presence of interspersed normal cells and 6 mitotic figures/10 HPF, favoring a diagnosis of bizarre nuclei-leiomyomas. For the remaining 5/6 cases the degree of atypia was estimated high by the pathologist and low by the reviewers. For 3/8 patients with recurrent disease we could analyze the second tumor, whose morphology led to a clear categorization of the tumor as a leiomyosarcomas (Figure 1b).
Morphological features of some challenging uterine smooth muscle tumors. (a) Case 17: cellular smooth muscle tumor with mild atypia and 20 mitotic figures (arrow). (b) This patient developed a pelvic and para-aortic recurrences 36 and 48 months later. The atypia is more prominent and mitotic count is higher. (c) Case 21: highly cellular smooth muscle tumor composed of monotonous atypical spindle cells with brisk mitotic activity. The patient is alive with disease at 156 months after two recurrences. (d) Case 25: cellular spindle cell tumor with diffuse mild to moderate atypia and seven mitotic figures. The patient developed bone metastases 10 months after the diagnosis and died of the disease 16 months later.
The average age was 50 years (range 24–85) and the median tumor size 7.7 cm. Out of 29, 22 patients underwent hysterectomy (for case 28 a curetting was followed by hysterectomy), 6/29 patients had myomectomy and for 1/29 patient (case 24) the surgical procedure was not specified. All tumors were stage I at presentation except one that was stage II (case 13) and another one (case 21) for which the status was unknown. For 27 tumors the quality of the surgery was optimal (R0), for one case suboptimal (R1) and for the remaining case the data was unknown. The follow-up period ranged from 12 to 156 months (overall average 70 months; 70 months in patients with myomectomy and 39 months for patients with hysterectomy) and details were obtained for all patients but one (case 7). For the leiomyomas control group follow-up details were obtained for 8/10 patients and all were alive and free of the disease. Follow-up was available for 8/10 patients of the leiomyosarcomas control group (6/8 patients died because of the disease and 2/8 were alive with disease).
Genomic Data and Clinical Correlation
The reference group of leiomyomas presented a flat genomic profile with no or only very-few sporadic alterations (Figure 2a). The group of leiomyosarcomas, on the other hand, showed a rearranged chromosome profile with numerous intrachromosomal breaks (Figure 2b). Deletions occurred more often than gains, the most frequent ones being 22q (70%), 13q13-14 (50%), 11p14-15 (50%), 6q12-16 (50%), and 3q25 (50%) and 1q21-22 (60%), respectively.
The two groups of tumors were therefore clearly distinct from each other as the genomic index assessment showed that all the leiomyomas harbored a genomic index <10 (between 0 and 7) and all leiomyosarcomas showed a genomic index >10 (between 19.6 and 80). The genomic study mirrored perfectly the morphological analysis and stratified the tumors in a benign group (leiomyomas) with a genomic index <10, and in a malignant group (leiomyosarcomas), involving a significant rate of metastases and unfavorable outcome, and with a genomic index >10.
Among the patients of the leiomyomas group, 7/10 were alive without evidence of the disease, one patient was alive with breast cancer and 2/10 were lost at follow-up. Among the patients with leiomyosarcomas, 6/10 were dead of the disease, 2/10 alive with disease and 2/10 lost at follow-up.
Upon establishing the threshold for malignancy at genomic index=10, the genomic analysis was applied to STUMP tumors.
The STUMP analysis showed a more heterogenous group of genomic profiles since we observed tumors with an even profile (Figure 3a), as well as those with a rearranged profile (Figure 3b). The latter harbored mainly the same alterations as leiomyosarcomas except for gains that were more frequent in leiomyosarcomas than in STUMP, and losses at 3q25, 6q12-16, and 22q that were less frequent in rearranged STUMP.
In 16/29 cases, the genomic index was <10 (Table 1). For this group of tumors, the follow-up was available in 15/16 cases and all patients were alive without evidence of the disease after an average time of 80 months (from 12 to 137 months). Two cases showing some worrisome morphological features such as diffuse atypia (case 14) and tumor necrosis (case 16) displayed a relatively flat, ‘leiomyoma-type’ benign profile in the genomic analysis. In this group of tumors no recurrence was recorded.
For the group of 13/29 tumors with genomic index>10, 3/13 patients were dead of the disease at 16, 30, and 64 months after the diagnosis and the recurrences appeared at 8, 10, and 56 months from the first diagnosis. Three out of thirteen patients were alive with disease with a follow-up of 60, 89, and 156 months. For 2/3, the recurrences were multiple in time and location (Table 1). One patient was alive, not otherwise specified (with 25 months of follow-up) and 6/13 patients were alive without evidence of the disease with an average time of 60 months (from 16 to 156 months). Of note, in the group of patients alive without evidence of the disease with genomic index>10, one patient (case 17) suffered two recurrences: one at 36 months from diagnosis and the second 12.4 months after the first recurrence. The alive not otherwise specified patient also had a recurrence 17 months after the first diagnosis.
The stratification according to the genomic index=10 threshold splits the STUMP category of tumors into two groups with different outcomes and a P-value not significant for overall survival and significant (P=0.00029, HR [95% IC] 3.2.10e9 [0-inf]) for metastasis-free survival (Table 2, Figure 4a). When comparing leiomyoma, leiomyosarcomas, and STUMP groups we observed that, although the STUMP with genomic index<10 showed the same clinical outcome as leiomyomas, those with genomic index >10 behaved like leiomyosarcomas (Figure 4b and c).
Kaplan–Meier analysis of metastasis-free survival for STUMP (a) and metastasis-free survival for STUMP, leiomyomas, and leiomyosarcomas (b) according to GI. Kaplan–Meier analysis of overall survival (c) of STUMP split according to GI compared with leiomyomas and LMS. GI, genomic index; GI1, genomic index<10; GI2, genomic index>10; MFS, metastasis-free survival; OS, overall survival.
We tested the prognostic value of atypia and found that it was statistically significant for metastasis-free survival (P=0.00060, HR[IC 95%] 15.82[1.93–129.74]) but not for overall survival. The presence of mitosis (cutoff of 10) and necrosis were not significant for either overall survival or metastasis-free survival (Table 2).
By multivariate analysis, genomic index was the only independent prognosis factor for metastasis-free survival (P=0.036, HR [IC 95%] 17.4 [1.17–2509]) (Table 2).
Discussion
The histological distinction between benign and malignant uterine smooth muscle lesions based on the ‘three features’ histological approach derived from the Stanford study2 and approved in 2014 by the WHO3 is generally straightforward. Nevertheless, in certain cases, a degree of subjectivity in the interpretation of these criteria, and the interposition of external factors such as prior treatments (hormonal therapy or embolization before surgery), or a nonoptimal fixation of the specimen, may pose challenges that could lead to an equivocal diagnosis. Some lesions with mild atypia and a benign appearance could, for instance, surprise with an unexpected recurrence or metastasis.18 Some leiomyosarcomas cases with confirmed histological malignancy, on the other hand, could end up having a prolonged survival even if the overall prognosis for leiomyosarcomas is poor.19, 20 The pathologist’s interpretation can also be particularly difficult and subjective, especially for patients who have ischemic, myxoid, or epithelioid modifications in leiomyoma caused by previously received endocrine therapy. Furthermore, an important interobserver variability in the interpretation of tumor cell necrosis in smooth mesenchymal uterine lesions was recently reported, with complete disagreement being reported in 18% of the cases.21 According to the 2014 WHO classification3 ‘a smooth muscle tumor with features that preclude an unequivocal diagnosis of leiomyosarcoma, but that do not fulfill the criteria for leiomyoma, or its variants, and raise concern that the neoplasm may behave in a malignant fashion should be termed ‘smooth muscle tumor of uncertain malignant potential’. Furthermore, STUMP represents a ‘managerial’ category rather than a biological and clinical entity, and for this reason the use of this diagnostic term should be limited whenever possible. This borderline category for which malignancy cannot be confirmed and the prognosis cannot be determined, represents a major diagnostic challenge that could have important clinical consequences for the treatment, as well as a profound human and social effect for the patient.
Our hypothesis is that the clinical behavior of smooth muscle uterine tumors is strongly associated with specific genomic profiles. In this study, we analyzed by array-CGH the genomic profile of a series of 29 problematic uterine smooth muscle lesions for which the assessment of the mitotic count, or of the presence and the degree of atypia, or of the nature of the necrosis were not conclusive and for which there was a poor interobserver agreement.
To test the pertinence of the genomic profile analysis, we first proceeded to the study of benign (leiomyoma) and malignant lesions (leiomyosarcomas) for which there was a complete diagnostic agreement. We observed a perfect concordance between morphology, clinical behavior, and genomic index. Genomic index was then applied to split the series of problematic smooth muscle uterine tumors into two groups: the first one (genomic index <10) without recurrence or metastasis and similar behavior to that of leiomyoma; and the second group (genomic index >10) consisting of tumors with a significant recurrence because of the disease that encompasses a spectrum of smooth muscle lesions approaching the recurrence risk level of leiomyosarcomas but a more indolent, less aggressive course compared with leiomyosarcomas (Figure 4). These data make the genomic index an independent prognostic marker in uterine mesenchymal lesions in terms of metastasis-free survival. The nonsignificant results in term of overall survival could be explained by the fact that there are more metastatic recurrence events in the cohort than deaths. A longer period of follow-up and a larger series could improve the prognostic power of genomic index for overall survival. Furthermore, it is judicious to stress that, given the limited period of follow-up for some tumors in our study, a potentially late, indolent recurrence might have gone undetected. Among the morphological parameters of Stanford criteria, the atypia is closest to GI as prognostic prediction. However, the presence of atypia alone does not allow the diagnosis of leiomyosarcomas.2 Also, mitotic activity is not a good single diagnostic parameter since it could reflect a reactive process. Furthermore, the mitotic count could be difficult to assess, especially in the ischemic areas where smudged nuclei in cellular degeneration or apoptosis may be confused with true mitotic figures causing an important interobserver variability. It is important to emphasize that we limited this exploratory study to conventional (spindle cell) uterine smooth muscle lesions, excluding those with any epithelioid or myxoid or bizarre nuclei component.
In the genomic era, few studies in the literature have reported on leiomyoma, leiomyosarcomas, and array-CGH, and only for a limited number of cases. In 2005, Cho et al13 studied the genomic profile of four leiomyoma and seven leiomyosarcomas by array-CGH with 1440 human BACs. Although uterine leiomyomas did not present genomic alterations, all leiomyosarcomass showed gains of 7q and 12q and several chromosome region losses. Although this study highlights the genomic differences between leiomyoma and leiomyosarcomas, supporting our hypothesis, the authors did not carry out the genome profiling as a practical tool to improve the morphological diagnosis. Meadows et al14 have recently studied loss of heterozygosity and copy number amplifications by high-density SNP platform (Affymetrix 100 K) in 37 leiomyomas. They observed that genome-wide loss of heterozygosity and copy number alterations are infrequent events in leiomyomas that generally do not have a clinical impact, confirming our hypothesis. They did not compare the leiomyomas with the leiomyosarcomas profile. Lastly, Raish et al15 described the main genetic alterations in 15 leiomyosarcomas.
To our knowledge, this is the first report on the use of genomic profile analysis by array-CGH as a diagnostic tool and of a genomic index threshold (<10 and >10) as a criterion for malignancy.
The comparison of different studies on the clinical behavior of STUMP is difficult because of differing and evolving diagnostic criteria and the variable terminology, as often in literature ‘atypical leiomyomas’ was employed as a synonym of STUMP as well as bizarre nuclei leiomyomas,12 a benign variant of leiomyomas.22 Elsewhere, the investigators from Stanford did not use the term ‘STUMP’, but instead four categories of uterine smooth muscle tumors that had a low or uncertain malignant potential: atypical leiomyomas with limited experience; smooth muscle tumor with low malignant potential; atypical leiomyomas with low risk of recurrence; and mitotically active leiomyomas with a limited experience.2 A meta-analysis performed on all published studies that can be grouped into the similar diagnostic criteria, with a sufficient follow-up and pathological details, reveals a recurrence rate ranging from 10.4 to 26.7%.3 Regardless of the ‘virtual’ category of STUMP, the genomic profile analysis is a pertinent and useful tool assisting the diagnosis in challenging smooth muscle uterine lesions.
In contrast to ‘high-grade’ leiomyosarcomas, recurrent STUMP are in general biologically low-grade tumors, as the recurrence is often delayed and the clinical course prolonged.12 These data support our findings.
As described above, the genomic index divides problematic uterine smooth muscle lesions into leiomyomas and leiomyosarcomas. However, this enlarges the range of leiomyosarcomas adding more indolent cases to conventional leiomyosarcomas, which have a more aggressive clinical course. In conclusion, we have demonstrated in this series that genomic index is a powerful tool that allow splitting the problematic uterine smooth muscle lesions in two categories according to the risk of recurrence: a group of benign tumors, similar to leiomyomas, and a group of malignant lesions, similar to leiomyosarcomas. Therefore, the genomic analysis by array-CGH is an innovative diagnostic tool for smooth muscle uterine tumors, complementary to the morphological approach, and it is suitable for differentiating between benign (leiomyomas) and malignant tumors (leiomyosarcomas) and for providing an improved classification of the intermediate category with uncertain malignancy (STUMP) in cases with equivocal morphological features.
References
Abeler VM, Royne O, Thoresen S et al. Uterine sarcomas in Norway. A histopathological and prognostic survey of a total population from 1970 to 2000 including 419 patients. Histopathology 2009;54:355–364.
Bell SW, Kempson RL, Hendrickson MR . Problematic uterine smooth muscle neoplasms. A clinicopathologic study of 213 cases. Am J Surg Pathol 1994;18:535–558.
Oliva E, Carcangiu ML, Carinelli SG et al. Mesenchymal tumours. Smooth muscle tumour of uncertain malignant potential In: Kurman RJ, Carcangiu ML, Herrington CS, Young RH, eds. WHO Classification of Tumours of Female Reproductive Organs. IARC: Lyon, 2014, pp 135–147.
de Vos S, Wilczynski SP, Fleischhacker M et al. p53 alterations in uterine leiomyosarcomas versus leiomyomas. Gynecol Oncol 1994;54:205–208.
Hall KL, Teneriello MG, Taylor RR et al. Analysis of Ki-ras, p53, and MDM2 genes in uterine leiomyomas and leiomyosarcomas. Gynecol Oncol 1997;65:330–335.
Jeffers MD, Farquharson MA, Richmond JA et al. p53 Immunoreactivity and mutation of the p53 gene in smooth muscle tumours of the uterine corpus. J Pathol 1995;177:65–70.
Mayerhofer K, Lozanov P, Bodner K et al. Ki-67 expression in patients with uterine leiomyomas, uterine smooth muscle tumors of uncertain malignant potential (STUMP) and uterine leiomyosarcomas (LMS). Acta Obstet Gynecol Scand 2004;83:1085–1088.
Niemann TH, Raab SS, Lenel JC et al. p53 protein overexpression in smooth muscle tumors of the uterus. Hum Pathol 1995;26:375–379.
O'Neill CJ, McBride HA, Connolly LE et al. Uterine leiomyosarcomas are characterized by high p16, p53 and MIB1 expression in comparison with usual leiomyomas, leiomyoma variants and smooth muscle tumours of uncertain malignant potential. Histopathology 2007;50:851–858.
Ip PP, Cheung AN, Clement PB . Uterine smooth muscle tumors of uncertain malignant potential (STUMP): a clinicopathologic analysis of 16 cases. Am J Surg Pathol 2009;33:992–1005.
Ip PP, Tse KY, Tam KF . Uterine smooth muscle tumors other than the ordinary leiomyomas and leiomyosarcomas: a review of selected variants with emphasis on recent advances and unusual morphology that may cause concern for malignancy. Adv Anat Pathol 2010;17:91–112.
Ip PP, Cheung AN . Pathology of uterine leiomyosarcomas and smooth muscle tumours of uncertain malignant potential. Best Pract Res Clin Obstet Gynaecol 2011;25:691–704.
Cho YL, Bae S, Koo MS et al. Array comparative genomic hybridization analysis of uterine leiomyosarcoma. Gynecol Oncol 2005;99:545–551.
Meadows KL, Andrews DM, Xu Z et al. Genome-wide analysis of loss of heterozygosity and copy number amplification in uterine leiomyomas using the 100 K single nucleotide polymorphism array. Exp Mol Pathol 2011;91:434–439.
Raish M, Khurshid M, Ansari MA et al. Analysis of molecular cytogenetic alterations in uterine leiomyosarcoma by array-based comparative genomic hybridization. J Cancer Res Clin Oncol 2012;138:1173–1186.
Chibon F, Lagarde P, Salas S et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med 2010;16:781–787.
Lagarde P, Perot G, Kauffmann A et al. Mitotic checkpoints and chromosome instability are strong predictors of clinical outcome in gastrointestinal stromal tumors. Clin Cancer Res 2012;18:826–838.
Amant F, Moerman P, Vergote I . Report of an unusual problematic uterine smooth muscle neoplasm, emphasizing the prognostic importance of coagulative tumor cell necrosis. Int J Gynecol Cancer 2005;15:1210–1212.
D'Angelo E, Espinosa I, Ali R et al. Uterine leiomyosarcomas: tumor size, mitotic index, and biomarkers Ki67, and Bcl-2 identify two groups with different prognosis. Gynecol Oncol 2011;121:328–333.
Veras E, Zivanovic O, Jacks L et al. ‘Low-grade leiomyosarcoma’ and late-recurring smooth muscle tumors of the uterus: a heterogenous collection of frequently misdiagnosed tumors associated with an overall favorable prognosis relative to conventional uterine leiomyosarcomas. Am J Surg Pathol 2011;35:1626–1637.
Lim D, Alvarez T, Nucci MR et al. Interobserver variability in the interpretation of tumor cell necrosis in uterine leiomyosarcoma. Am J Surg Pathol 2013;37:650–658.
Croce S, Young RH, Oliva E . Uterine leiomyomas with bizarre nuclei: a clinicopathologic study of 59 cases. Am J Surg Pathol 2014;38:1330–1339.
Acknowledgements
We thank the Ligue contre le Cancer, Fondation ARC and EUROSARC for funding. FA is senior clinical researcher and TC is researcher for the Research Fund Flanders (FWO). FA, TC, and MZ are members of ENITEC (European Network on Individualized Treatment in Endometrial Cancer). We thank Jone Iriondo-Alberdi for medical writing assistance in English.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Author contributions
JC Noel, M Devouassoux-Shisheborah, F Amant, T Cuppens, L Arnould, F Mishellany, M Zikan, A Leroux, E Frouin, P Duvillard, P Terrier, I Farre, I Valo, E Stoekle, A Floquet, F Guyon and D Garbay supplied tumor tissue and collected clinical follow-up data; A Ribeiro carried out laboratory experiments; F Chibon supervised and interpreted laboratory experiments; C Brulard conducted statistical analysis; S Croce and G Macgrogan collected tumor sample and did the central pathological review; and S Croce and F Chibon designed the study and wrote the report. All the contributor authors reviewed and approved the final report.
Rights and permissions
About this article
Cite this article
Croce, S., Ribeiro, A., Brulard, C. et al. Uterine smooth muscle tumor analysis by comparative genomic hybridization: a useful diagnostic tool in challenging lesions. Mod Pathol 28, 1001–1010 (2015). https://doi.org/10.1038/modpathol.2015.3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2015.3
This article is cited by
-
Molecular and clinicopathologic characterization of intravenous leiomyomatosis
Modern Pathology (2020)
-
Uterine and vaginal sarcomas resembling fibrosarcoma: a clinicopathological and molecular analysis of 13 cases showing common NTRK-rearrangements and the description of a COL1A1-PDGFB fusion novel to uterine neoplasms
Modern Pathology (2019)
-
Myxoid smooth muscle neoplasia of the uterus: comprehensive analysis by next-generation sequencing and nucleic acid hybridization
Modern Pathology (2019)
-
Myome – was liegt da in der DNA?
Der Gynäkologe (2019)
-
Genome profiling is an efficient tool to avoid the STUMP classification of uterine smooth muscle lesions: a comprehensive array-genomic hybridization analysis of 77 tumors
Modern Pathology (2018)