Abstract
Sequencing of clear cell renal cell carcinomas identified loss-of-function mutations of SETD2, a gene that encodes a nonredundant methytransferase responsible for histone H3 lysine 36 trimethylation (H3K36me3), and H3K36me3 is progressively deregulated in metastases. However, few data exist regarding the impact of loss of H3K36me3 on outcomes. We assessed the association of SETD2 DNA alterations and mRNA expression with overall survival using The Cancer Genome Atlas clear cell renal carcinoma data (N=411). Additionally, we assessed the association of H3K36 loss of methylation with renal cell carcinoma-specific survival and progression-free survival using an independent cohort at Mayo Clinic (N=1454). Overall survival, renal cell carcinoma-specific survival and progression-free survival were estimated using Kaplan–Meier method, and differences in survival across groups was compared using Cox regression models, adjusted for age and the Mayo SSIGN (stage, size, grade, and necrosis) score. In The Cancer Genome Atlas cohort, SETD2 DNA alterations or mRNA expression was not associated with overall survival (P>0.05). In the Mayo cohort, patients with H3K36me3-negative tumors were two times more likely to experience renal cell carcinoma-specific death than patients with H3K36me3-positive tumors (hazard ratio, 2.23; 95% confidence interval, 1.77–2.81); P<0.0001. After stratifying for the SSIGN score, H3K36me3-negative tumors in the low-risk SSIGN group had a worse renal cell carcinoma-specific survival (hazard ratio, 2.18; 95% confidence interval, 1.09–4.36); P=0.03. Although SETD2 DNA and mRNA alterations are not associated with overall survival, we provide evidence that deregulation of the H3K36me3 axis is associated with a higher risk of renal cell carcinoma-specific death. This association remains significant after stratifying for the SSIGN score, particularly among those patients with low-risk tumors.
Similar content being viewed by others
Main
The molecular pathogenesis of clear cell renal cell carcinoma is linked to chromosome arm 3p copy number loss and biallelic inactivation of the von Hippel-Lindau gene located on chromosome 3p. More recently, mutations in other tumor suppressors on chromosome 3p such as PBRM1 (~40%), BAP1 (~10%), and SETD2 (~12%) have been identified as common events in clear cell renal cell carcinoma tumors.1 To date, however, there are no investigations that have evaluated associations between loss of SETD2 function and clear cell renal cell carcinoma outcome.
The SETD2 gene encodes a nonredundant histone 3 lysine 36 trimethyltransferase and is mutated in other cancers.2 In Setd2-knockout mice, histone 3 lysine 36 trimethylation (H3K36me3) is absent,3 and in humans, loss-of-function SETD2 mutations are associated with loss of H3K36me3.4, 5, 6 Of note, sequencing of various human tumors identified recurrent molecular alterations that phenotypically converge on deregulation of the H3K36me3 axis, and H3K36me3 is progressively deregulated in clear cell renal cell carcinoma metastases.7 Motivated by this and the aforementioned lack of data on H3K36me3 loss and clear cell renal cell carcinoma outcome, we employed our own immunohistochemistry-based assay for H3K36me3 in archival formalin-fixed, paraffin-embedded tissue sections for which negative staining correlates with a SETD2 mutant genotype.6 We hypothesized that disruption of the histone code at H3K36me3 is associated with an increased risk of cancer-specific death. Moreover, we explore the deeper clinical relevance of this association by evaluating whether loss of H3K36me3 is associated with outcome among the specific subset of clear cell renal cell carcinoma patients already determined to have ‘low-risk’ disease based on the externally validated Mayo Clinic SSIGN (stage, size, grade, and necrosis) prognostic scoring system.8, 9
Materials and methods
Patient Selection
After Mayo Clinic Institutional Review Board approval, we identified 1465 patients treated with radical nephrectomy or nephron-sparing surgery for clear cell renal cell carcinoma between 1990 and 2009 from the Mayo Clinic Nephrectomy Registry with representative paraffin-embedded tissue blocks available for immunohistochemistry staining and data on renal cell carcinoma-specific death. After review of the entire case, one representative slide was selected with the highest Fuhrman grade and tumor content for immunohistochemistry staining. A genitourinary pathologist (JC) reviewed all the tumors, which allowed for standardized clinicopathological variables.
Assessment of H3K36me3, PBRM1, and BAP1 by Immunohistochemistry Staining
Standard immunohistochemistry staining procedures for H3K36me3, PBRM1, and BAP1 were performed using the Dako (Carpinteria, USA) autostainer and Ventana (Tucson, USA) Benchmark XT automated stainer. After heat-induced epitope retrieval with Cell Conditioning Solution 1 (Ventana), sections were incubated with the appropriate primary antibody: H3K36me3 (Abcam 9050, Cambridge, USA) at 1:9000 (15 min); PBRM1 (Bethyl Laboratories A301–591 A, Montgomery, USA) at 1:250 (32 min), BAP1 (Santa Cruz Biotechnology sc-28383, Dallas, USA) at 1:50 (60 min). We previously validated immunohistochemistry assays to evaluate H3K36me3, PBRM1, and BAP1 protein expression in which negative staining correlated with loss-of-function mutations in SETD2, PBRM1, and BAP1 genes, respectively.6, 10 Samples were excluded from analysis if positive nuclear staining was not observed in background stromal cells or lymphocytes (internal control). Positivity (2+ staining intensity) was indicated by diffuse nuclear staining in tumor cells (≥10%); cytoplasmic staining was not analyzed. Samples with little-to-no tumor nuclei staining were classified as negative. Samples with positive nuclei in the internal control tissue (stroma and/or lymphocytes) and faint tumor nuclei staining were classified as weak positive (1+ staining intensity). Focal negatives had positive nuclei in the internal control tissue and had loss of tumor nuclear staining only in subclonal populations (<10% of total tumor nuclei). For the purposes of dichotomizing the H3K36me3 classifications (positive, negative, weak positive, focal negative), we classified weak positive as positive and focal negative as negative based on our immunohistochemistry results in the tumors with a defined SETD2 genotype. With respect to H3K36me3 classification, the genitourinary pathologists (PK and MLS) were blinded to all clinical outcomes and SETD2 genotypes.
Statistical Analyses
The Fisher exact or Chi-square tests, as appropriate, were used to compare categorical variables across molecular groups. Cox proportional hazards models and hazard ratio with 95% confidence interval were used to assess the association of H3K36me3, PBRM1, and BAP1 expression with outcome after adjusting for age and/or the Mayo Clinic SSIGN score. Overall survival was assessed in The Cancer Genome Atlas data, whereas renal cell carcinoma-specific survival and progression-free survival were assessed in the Mayo Clinic data. Renal cell carcinoma-specific survival analyses tracked time from nephrectomy to death owing to renal cell carcinoma, whereas progression-free survival considered either the first metastasis or renal cell carcinoma-death as an end point. In both analyses, patients who died from other causes or were lost to follow-up were censored at those respective times. We calculated concordance index values to assess predictive ability. All reported concordance indices were generated using the bootstrap methodology and represent optimism-corrected estimates of concordance.11 The Kaplan–Meier method was used to estimate time to renal cell carcinoma-specific death and progression-free survival. Statistical analyses were performed using R, version 2.15. For all analyses, a P-value <0.05 was evidence of statistical significance.
Results
Impact of SETD2 Expression on Overall Survival from The Cancer Genome Atlas Clear Cell Renal Cell Carcinoma Data Set
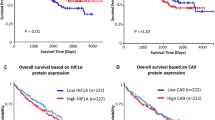
The impact of SETD2 DNA copy number loss or mRNA expression on overall survival is unknown. 3p loss of heterozygosity occurs in >90% of clear cell renal cell carcinoma cases, and a loss-of-function SETD2 mutation in the remaining allele would lead to biallelic inactivation of SETD2. We assessed the impact of SETD2 copy number loss alone, concurrent SETD2 copy number loss and SETD2 mutation and no SETD2 copy number loss or mutations on overall survival (Figure 1a, Supplementary Figure S1). We did not observe an association (overall log-rank test P=0.25) between overall survival and SETD2 copy number alterations (hazard ratio=0.66, 95% confidence interval=0.37–1.18) or concurrent copy number loss with SETD2 mutation (hazard ratio=0.9, 95% confidence interval=0.44–1.82). To determine whether overall survival was associated with SETD2 mRNA expression, we dichotomized mRNA (high or low) expression at the median RNA-Sequencing by Expectation Maximization value (Figure 1b). As with SETD2 copy number alterations, we did not observe an association between high SETD2 mRNA expression and overall survival (low SETD2 expression hazard ratio (95% confidence interval), 1.28 (0.91–1.80) (P=0.16, log-rank test)).
Kaplan–Meier survival plots for SETD2 DNA and RNA in The Cancer Genome Atlas kidney renal clear cell carcinoma data set. (a) Analysis of SETD2 copy number and mutations. (b) Analysis of SETD2 mRNA dichotomized as high or low expression based on median RNA-Sequencing by Expectation-Maximization value. Two samples were missing for overall survival.
Correlation of Mutant SETD2 Genotype with Loss of H3K36me3 Phenotype
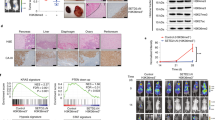
Setd2-knockout mice lack global H3K36me3,3 and in humans, loss-of-function SETD2 mutations in various tumors are associated with loss of H3K36me3.3, 5, 6 To evaluate H3K36me3 expression as a dichotomized variable using an immunohistochemistry assay in which loss of H3K36me3 correlates with SETD2 mutations, we generated targeted SETD2 deletions in the 786-O renal cell carcinoma cell line using zinc-finger nucleases. In a clone, 786-O SETD2 zinc finger nuclease, a four-base deletion in SETD2 was confirmed by Sanger sequencing (Supplementary Figure S2). Consistent with our prior study in an independent SETD2 zinc-finger nuclease clone, SETD2 deletion disrupts the expression of H3K36me3 by western blotting and immunohistochemistry assays (Figures 2a and b).7 To validate H3K36me3 expression as a dichotomized variable, H3K36me3 was scored by two pathologists (PK and MLS), blinded to the SETD2 genotype. Of the 26 SETD2-genotyped tumors, 21 (81%) were classified as H3K36me3 positive or negative, 4 as focal negative, and 1 as weak positive (Supplementary Table S1, Figures 2c–f). Of the 15 tumors with a SETD2 wild-type genotype, 12 were classified as positive, 2 as focal negative, and 1 as weak positive. Of the 11 tumors with a SETD2 mutant genotype, 9 were classified as negative and 2 as focal negative. Overall, 81% of the tumors using immunohistochemistry classifications of negative or positive correlated with the known SETD2 genotype; the SETD2 genotype–H3K36me3 phenotype concordance improves to 100% after exclusion of the five heterogeneous staining tumors classified as weak positive or focal negative.
Analysis of H3K36me3 in isogenic SETD2 renal cell carcinoma cell lines and The Mayo Clinic Nephrectomy Registry. The SETD2 wild-type 786-O cell line was transfected with zinc finger pairs that generate a SETD2 deletion. Single cell-derived clones were analyzed by fragment length analysis to identify clone SETD2 zinc finger nuclease. SETD2 was sequenced and confirmed to have a four-base deletion. (a) Western blotting confirming depletion of H3K36me3 with histone H3 and actin as controls. (b) Immunohistochemical staining of H3K36me3 comparing isogenic SETD2 cell lines. Representative images for H3K36me3 immunohistochemical staining classifications: (c) positive, (d) negative, (e) weak positive, and (f) focal negative in nephrectomy samples. Scale bar 50 μm (original magnification 400 × ; inset 1000 ×). Kaplan–Meier estimate of (g) renal cell carcinoma-specific death and (h) progression-free survival in patients with H3K36me3-negative and -positive tumors. H3K36me3 indicates histone 3 lysine 36 trimethylation.
Association of H3K36me3 with Pathological Characteristics, Renal Cell Carcinoma-Specific Death, and Progression-Free Survival
Of the initial 1465 slides, 1454 (99.2%) had successful immunohistochemistry staining for H3K36me3. The 1454 slides were stratified into two categories of H3K36me3 results: positive (1066; 73.3%) or negative (388; 26.7%) (Supplementary Figure S3). Tumors classified as H3K36me3 negative were associated with larger tumor size, higher grade, and increased tumor necrosis (Table 1). Our mean duration of follow-up is 8.1 years (median, 8.3 years; range, 0–23.5 years; with 6 patients missing follow-up).
When H3K36me3 was modeled as a dichotomous variable, we observed evidence of an increase in risk of renal cell carcinoma-specific death and progression-free survival with loss of H3K36me3. In Figures 2g and h after adjusting for age, patients with H3K36me3-negative tumors were two times more likely to experience renal cell carcinoma-specific death and progression than patients with H3K36me3-positive tumors (hazard ratio (95% confidence interval), 2.23 (1.77–2.81); P<0.0001 and hazard ratio (95% confidence interval), 2.12 (1.74–2.60); P<0.0001, respectively). After excluding those with heterogeneous staining (weak positive, focal negative), patients with H3K36me3-negative tumors were approximately three times more likely to experience renal cell carcinoma-specific death and progression (Supplementary Table S2). As loss of H3K36me3 is associated with greater tumor size, grade, and necrosis, we evaluated the prognostic value of H3K36me3 expression after adjusting for the SSIGN score, an externally validated prognostic scoring system (Supplementary Table S3). Adjustment for the SSIGN score results in attenuation of the association of H3K36me3 with risk of renal cell carcinoma-specific death (hazard ratio (95% confidence interval), 1.26 (0.99–1.60); P=0.06). Subsequently we stratified the tumors by SSIGN groups (0–3, 4–7, and 8+) into low-, intermediate-, and high-risk groups, respectively. In the high-risk group (SSIGN 8+), we did not observe an association between H3K36me3 expression and renal cell carcinoma-specific survival (Table 2). However, H3K36me3-negative tumors in the low- and intermediate-risk SSIGN groups had a worse renal cell carcinoma-specific survival and progression-free survival. In Figure 3, we provide the stratified Kaplan–Meier curves for the low-, intermediate-, and high-risk SSIGN groups. Exclusion of those with heterogeneous staining (Table 3) improves upon the prognostic ability; patients with H3K36me3-negative tumors in the low- and intermediate-risk SSIGN were more likely to experience renal cell carcinoma-specific death (hazard ratio (95% confidence interval), 2.18 (1.09–4.36); P=0.03 and hazard ratio (95% confidence interval), 1.45 (1.01–2.07); P=0.04. Our data indicate that loss of H3K36me3 expression is associated with a higher risk of renal cell carcinoma-specific death and progression; this association remains significant after correcting for age and SSIGN score.
Kaplan–Meier Estimate of renal cell carcinoma-specific death and progression-free survival in patients with H3K36me3 negative and positive tumors by low-, intermediate-, or high-risk Mayo SSIGN scores. (a) Renal cell carcinoma-specific survival. (b) Progression-free survival. H3K36me3 indicates histone 3 lysine 36 trimethylation; SSIGN, stage, size, grade, and necrosis.
Association of H3K36me3 with Epigenetic Tumor Suppressors BAP1 and PBRM1
The molecular pathogenesis of clear cell renal cell carcinoma is linked to chromosome 3p copy number loss and subsequent biallelic inactivation of the 3p tumor-suppressors VHL, SETD2, BAP1, and PBRM1. We previously evaluated the prognostic impact of BAP1 and PBRM1 to define epigenetic subtypes of clear cell renal cell carcinoma.12, 13 To examine the association of H3K36me3 with BAP1 and PBRM1 protein expression, we evaluated the expression of all three markers dichotomized as positive vs negative in clear cell renal cell carcinoma samples with available staining (Supplementary Figure S3). H3K36me3-negative tumors were almost three times more likely to be PBRM1-negative than were H3K36me3-positive tumors (odds ratio (95% confidence interval), 2.98 (2.14–4.17); P<0.0001; Supplementary Figure S4). We did not observe an association between H3K36me3 and BAP1 expression (P=0.31).
Discussion
Although recurrent DNA alterations in 3p tumor suppressors occur in The Cancer Genome Atlas clear cell renal cell carcinoma cohort, the prognostic impact of these alterations is limited after adjusting for the SSIGN score.14 Systematic molecular profiling of tumors such as chondrosarcomas, glioblastomas, and leukemias identified recurrent molecular alterations that phenotypically converge on functional deregulation of the H3K36me3 axis.4, 5, 15 Related to this, multi-region sequencing of clear cell renal cell carcinoma tumors has identified parallel evolution of distinct SETD2 mutations that phenotypically converge on loss of methyltransferase activity.16 In this study, we evaluated deregulation of the H3K36me3 axis using an immunohistochemistry assay in which loss of expression correlates with a SETD2-mutant genotype in cell lines and human clear cell renal cell carcinoma tumors. Our notable observations include: (1) alterations of SETD2 DNA and mRNA expression in clear cell renal cell carcinoma tumors from The Cancer Genome Atlas clear cell renal cell carcinoma cohort are not associated with overall survival; (2) loss of H3K36me3, a posttranslational histone modification, is associated with worse outcomes (renal cell carcinoma-specific survival, progression free survival); (3) the association with progression free survival remained significant after adjusting for age and SSIGN score; and (4) these associations remain apparent among the subset of clear cell renal cell carcinoma patients already determined to be in the intermediate-risk group based on the Mayo SSIGN score.
The association of H3K36me3-negative tumors with greater size, grade, and necrosis suggests a link between loss of H3K36me3 and renal cell carcinoma tumor progression. H3K36me3 regulates DNA repair, alternative splicing, and chromatin remodeling, and these functions are linked to chromatin ‘readers’ with proline–tryptophan–tryptophan–proline domains that interact with methylated lysine residues.17, 18, 19 Taken together, evidence supports a model in which loss of H3K36me3 or SETD2 ‘writer’ function may alter the cancer phenotype through deregulation of these chromatin ‘readers’. There are likely other posttranslational mechanisms that downregulate H3K36me3. In gliomas and chondroblastomas, mutations at lysine 36 in the histone H3.3 variant H3F3A are also associated with loss of H3K36me3.15, 20 We hypothesize that deregulation of the H3K36me3 axis may have a role in progression in other tumors.
There are limitations to this investigation that should be considered when evaluating our observations regarding loss of H3K36me3 and clear cell renal cell carcinoma outcome. First, we did not assess tumor genotype by massive parallel sequencing of all tumors in our cohort. Instead, we focused on a mechanistically relevant end point of loss of H3K36me3 expression after validation in cell line and tumors with a defined SETD2 genotype.7 DNA sequencing can detect single-nucleotide variants and allelic fractions; however, in the absence of germline or normal controls, germline polymorphisms can confound mutation calls. Furthermore, it was estimated in a sequencing study of multiple cores from primary renal cell carcinoma tumors that a minimum of three distinct cores are required for accurate tumor genotyping.21 In contrast, immunohistochemistry may be used to screen for functional loss of SETD2 without concurrent evaluation of germline DNA. Second, challenges exist in how to interpret the identification of tumors with heterogeneous staining (focal negative, weak positive). In our training cohort after exclusion of five tumors with heterogeneous staining, there is a 100% concordance between H3K36me3-positive/negative and SETD2 wild-type/mutant genotypes. On the basis of our immunohistochemistry training cohort of SETD2-genotyped tumors, two focal-negative tumors were SETD2 wild-type and two were SETD2 mutant; one SETD2 wild-type tumor was weak positive. The inclusion of heterogeneous staining samples in the training cohort reduces the concordance to 81% and attenuated the prognostic impact of H3K36me3 immunohistochemistry in the Nephrectomy Registry cohort; however, the association between H3K36me3 immunohistochemistry-negative tumors and worse outcomes remained significant.
Notwithstanding the above limitations, our work further extends the emerging molecular classification of clear cell renal cell carcinoma. Gerlinger et al22 have shown that intragenic VHL mutations and loss of 3p are the only uniformly truncal events in ccRCC. We have shown that loss of PBRM1 and BAP1 are mutually exclusive and that loss of PBRM1 is associated with better outcomes than loss of BAP1.10, 13, 23 Herein we validate previous results from a meta-analysis showing that PBRM1 and SETD2 mutations cooperate in ccRCC.24 Specifically, we find that H3K36me3 loss is 3 times more likely in PBRM1-negative tumors than in those that express PBRM1. These data further sub-classifies PBRM1-negative tumors into those with and without SETD2 loss.
Conclusions
In this study, we provide the first evidence that loss of H3K36me3 expression is associated with a higher risk of renal cell carcinoma-specific death and progression after nephrectomy for clinically localized clear cell renal cell carcinoma; this association remains significant after adjusting for age and SSIGN score. More importantly, the association remains apparent in the subset of patients with low-risk disease based on the Mayo SSIGN score, thus underscoring the potential for this biomarker to provide clinical utility to urologists. Indeed, if our results are confirmed in future studies, evaluation of H3K36me3 expression could be used alone or in combination with other biomarkers to improve risk stratification and clinical management of the growing number of patients presenting with low-risk clear cell renal cell carcinoma.
References
Creighton CJ, Morgan M, Gunaratne PH et al. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43–49.
Kudithipudi S, Jeltsch A . Role of somatic cancer mutations in human protein lysine methyltransferases. Biochim Biophys Acta 2014;1846:366–379.
Hu M, Sun XJ, Zhang YL et al. Histone H3 lysine 36 methyltransferase Hypb/Setd2 is required for embryonic vascular remodeling. Proc Natl Acad Sci USA 2010;107:2956–2961.
Mar BG, Bullinger LB, McLean KM et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun 2014;5:3469–3475.
Fontebasso AM, Schwartzentruber J, Khuong-Quang DA et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol 2013;125:659–669.
Simon JM, Hacker KE, Singh D et al. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res 2013;24:241–250.
Ho TH, Park IY, Zhao H et al. High-resolution profiling of histone H3 lysine 36 trimethylation in metastatic renal cell carcinoma. Oncogene 2015; doi:10.1038/onc.2015.221.
Zigeuner R, Hutterer G, Chromecki T et al. External validation of the Mayo Clinic stage, size, grade, and necrosis (SSIGN) score for clear-cell renal cell carcinoma in a single European centre applying routine pathology. Eur Urol 2010;57:102–109.
Frank I, Blute ML, Cheville JC et al. An outcome prediction model for patients with clear cell renal cell carcinoma treated with radical nephrectomy based on tumor stage, size, grade and necrosis: the SSIGN score. J Urol 2002;168:2395–2400.
Pena-Llopis S, Vega-Rubin-de-Celis S, Liao A et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet 2012;44:751–759.
Harrell FE Jr, Lee KL, Mark DB . Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 1996;15:361–387.
Kapur P, Pena-Llopis S, Christie A et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol 2013;14:159–167.
Joseph RW, Kapur P, Serie DJ et al. Loss of BAP1 protein expression is an independent marker of poor prognosis in patients with low-risk clear cell renal cell carcinoma. Cancer 2013;120:1059–1067.
Hakimi AA, Mano R, Ciriello G et al. Impact of recurrent copy number alterations and cancer gene mutations on the predictive accuracy of prognostic models in clear cell renal cell carcinoma. J Urol 2014;192:24–29.
Behjati S, Tarpey PS, Presneau N et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 2013;45:1479–1482.
Gerlinger M, Rowan AJ, Horswell S et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–892.
Vermeulen M, Eberl HC, Matarese F et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 2010;142:967–980.
Li F, Mao G, Tong D et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013;153:590–600.
Luco RF, Pan Q, Tominaga K et al. Regulation of alternative splicing by histone modifications. Science 2010;327:996–1000.
Bjerke L, Mackay A, Nandhabalan M et al. Histone H3.3. mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov 2013;3:512–519.
Sankin A, Hakimi AA, Mikkilineni N et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med 2014;3:1485–1492.
Gerlinger M, Horswell S, Larkin J et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 2014;46:225–233.
Joseph RW, Kapur P, Serie DJ et al. Clear cell renal cell carcinoma subtypes identified by BAP1 and PBRM1 expression. J Urol 2015; doi:10.1016/j.juro.2015.07.113.
Pena-Llopis S, Christie A, Xie XJ et al. Cooperation and antagonism among cancer genes: the renal cancer paradigm. Cancer Res 2013;73:4173–4179.
Acknowledgements
We thank the support provided by the Gloria A and Thomas J Dutson, Jr Kidney Research Endowment. This work was supported by the Mayo Clinic Center for Individualized Medicine Epigenomics Program (to THH); a Kathryn H and Roger Penske Career Development Award to Support Medical Research (to THH); Gerstner Family Career Development Award (to THH), the National Cancer Institute at the National Institutes of Health (K12CA90628 to THH, R01CA134466 to ASP, R01CA175754 to JB); Cancer Prevention Research Institute of Texas (RP130603 to JB). This publication was made possible by CTSA Grant Number UL1 TR000135 from the National Center for Advancing Translational Sciences, a component of the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Ho, T., Kapur, P., Joseph, R. et al. Loss of histone H3 lysine 36 trimethylation is associated with an increased risk of renal cell carcinoma-specific death. Mod Pathol 29, 34–42 (2016). https://doi.org/10.1038/modpathol.2015.123
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2015.123
This article is cited by
-
The role of histone methylation in renal cell cancer: an update
Molecular Biology Reports (2023)
-
Identification of DNA methylation signatures associated with poor outcome in lower-risk Stage, Size, Grade and Necrosis (SSIGN) score clear cell renal cell cancer
Clinical Epigenetics (2021)
-
H3K9me3, H3K36me3, and H4K20me3 Expression Correlates with Patient Outcome in Esophageal Squamous Cell Carcinoma as Epigenetic Markers
Digestive Diseases and Sciences (2019)
-
ATRX, DAXX or MEN1 mutant pancreatic neuroendocrine tumors are a distinct alpha-cell signature subgroup
Nature Communications (2018)
-
The spectrum of renal cell carcinoma in adults
Abdominal Radiology (2016)