Abstract
The aim of this study was to investigate the prevalence of fibroblast growth factor receptor 1 (FGFR1) amplification by fluorescence in situ hybridization (FISH) in a lung cancer patient cohort and to correlate results with morphology, silver in situ hybridization (SISH), and patient outcome. FGFR1 FISH and SISH were performed in 406 and 385 lung cancer cases, respectively, and the results were compared. High-level FGFR1 amplification was defined as the ratio of FGFR1/centromere 8 ≥2, or tumor cell percentage with ≥15 signals ≥10%, or average number of signals/tumor cell nucleus ≥6. Low-level amplification was defined as tumor cell percentage with ≥5 signals ≥50%. Of 406 tumors tested, there were 191 squamous cell carcinomas, 28 carcinomas with focal squamous morphology, 24 large cell carcinomas with squamous immunoprofile, 115 adenocarcinomas, 17 neuroendocrine tumors, and 31 carcinomas without squamous morphology or immunoprofile. FGFR1 FISH was assessable in 368 tumors, with FGFR1 amplification identified in 50, including 48 tumors with either squamous morphology or immunoprofile (48 of 225, 21.3%), and two ‘marker-null’ tumors without squamous or glandular morphology or immunoprofile (2 of 143, 1.4%; P<0.0001). FGFR1 SISH was assessable in 347 tumors. All 46 FGFR1 FISH-amplified tumors with tumor available for testing showed amplification with SISH, while all other tumors were negative. There was no relationship between FGFR1 amplification status and disease-free (P=0.88, HR=1.04, 95% confidence interval (CI)=0.67–1.60) or overall survival (P=0.97, HR=1.01, 95% CI=0.65–1.58) in surgically radically treated patients with tumors with any squamous morphology or immunoprofile. FGFR1 amplification is a common abnormality in tumors with any squamous morphology or immunoprofile, but it is also present in ‘marker-null’ tumors. The results of FGFR1 SISH showed 1:1 correlation with the results of FGFR1 FISH, indicating that SISH may be an alternative method to detect FGFR1 amplification. No relationship was detected between patient outcome and FGFR1 amplification.
Similar content being viewed by others
Main
In Australia in 2009, lung cancer was the fifth most commonly diagnosed cancer, but it was the most common cause of cancer deaths for both men and women.1 Approximately 85% of lung cancers are of non-small-cell carcinoma type, the subtypes of which are in reducing order of frequency: adenocarcinoma, squamous cell carcinoma, and large cell carcinoma, the latter being a morphologic diagnosis of exclusion. Major therapeutic advances have been made for patients with advanced lung adenocarcinoma owing to the discovery of molecular abnormalities for which there are effective targeted therapies.2, 3, 4, 5, 6, 7, 8 Discovery of actionable molecular targets in lung squamous cell carcinoma has lagged behind those made in adenocarcinoma.
A recent study has, however, identified a relatively common potentially targetable molecular abnormality in squamous cell carcinoma. In 2010, Weiss et al9 reported the presence of frequent and focal amplification of fibroblast growth factor receptor 1 (FGFR1) in 9.7% of lung squamous cell carcinoma cases by single-nucleotide polymorphism (SNP) arrays and in 22% of a separate cohort of lung squamous cell carcinoma cases by fluorescence in situ hybridization (FISH). In addition, testing of three separate cohorts of nonsquamous tumors identified FGFR1 amplification in 1 and 1.3% by SNP arrays and in 1% by FISH, respectively. Furthermore, Weiss et al demonstrated and validated the inhibition of growth and induction of apoptosis in separate FGFR1-amplified cell lines with the pan-FGFR inhibitor PD173074 or with short interfering RNA to FGFR1. They also demonstrated that FGFR1-amplified tumors showed significant tumor shrinkage with this small molecule in vivo. A later publication by Dutt et al10 reported FGFR1 amplification by SNP arrays in 21% of lung squamous cell carcinoma and 3.4% of lung adenocarcinoma (P<0.001), mostly in non-small cell lung carcinoma cell lines. This group also validated FGFR1 as a potential druggable target by demonstrating sensitivity of an FGFR1-amplified non-small cell lung carcinoma cell line with the pan-FGFR inhibitor PD173074.10
The FGFR1 gene, on chromosome 8, is one of the most frequently amplified genes in human cancer.11 The FGFR tyrosine kinase family is composed of four kinases, FGFR 1, 2, 3, and 4, acting as cell-surface receptors for fibroblast growth factors, each of which is critical in the control of embryonic development, cell proliferation, differentiation, and migration.12 This FGFR tyrosine kinase family also has an important role in tumor pathogenesis and can be dysregulated by amplification, point mutations, or translocation.13, 14 FGFR1 amplification has recently been reported by members of our group in oral squamous cell carcinoma,15 but it has also been described in esophageal squamous cell carcinoma, ovarian cancer, bladder cancer, prostate cancer, and rhabdomyosarcoma.16, 17, 18, 19, 20
Subsequent to the landmark initial studies,9, 10 other groups21, 22, 23, 24, 25, 26, 27 have investigated FGFR1 amplification in non-small cell lung carcinoma, using different definitions of FGFR1 amplification generally utilizing FISH, but there have been reports utilizing silver in situ hybridization (SISH)23 and real-time polymerase chain reaction.24
The frequency of FGFR1 amplification in squamous cell carcinoma and its potential as an actionable molecular target highlight the importance of not only defining amplification but also determining the most reliable and reproducible method to detect its presence, as well as assessing which tumor subtypes to test. These issues were addressed in 2012 by Schildhaus et al26 who proposed a standardized reading and evaluation strategy and evaluation criteria for FGFR1 FISH based on a study of 420 lung cancer patients. High-level FGFR1 amplification was defined as the ratio of FGFR1/centromere 8 (CEN8) ≥2, or the tumor cell percentage with ≥15 FGFR1 signals ≥10%, and the average number of FGFR1 signals/tumor cell nucleus ≥6, and low-level FGFR1 amplification as tumor cell percentage with ≥5 FGFR1 signals ≥50%. They reported FGFR1 amplification in 20% of squamous cell carcinomas (58 of 290), two of 13 ‘other’ tumors, and 0 of 97 adenocarcinomas (FGFR1/CEN8 ratio, t-test, P=0.01).
The aim of this study was to evaluate the prevalence of FGFR1 amplification by FISH in an Australian cohort of 406 lung cancers from 386 patients, using the reading and evaluation strategy and definitions proposed by Schildhaus et al, and to correlate these results with detailed morphologic and immunohistochemical assessment, and pathologic and clinical parameters, including overall survival and the results of SISH.
Patients and methods
Patients
A review of a prospectively maintained surgical database was used to identify patients with primary lung cancer resected from 1996 to 2012 at two tertiary institutions in Melbourne: St Vincent’s Hospital, Fitzroy, and Peter MacCallum Cancer Centre, East Melbourne. From this review, 388 tumors were identified and used to construct a tissue microarray. In addition, 18 tumors that underwent FGFR1 FISH testing on whole sections for clinical reasons, 11 of which were biopsies, were also included in the study. All 386 patients had a histopathologic diagnosis of primary lung cancer defined according to the 2004 World Health Organization (WHO) Classification of Lung Tumours.28 All tumors were staged according to the 7th edition tumor, node, metastasis (TNM) classification.29 Of the 406 tumors (from 386 patients), 368 tumors (from 352 patients) were able to be assessed by FISH. Clinical information was obtained from detailed prospective clinical databases. The definition of a never smoker was a person with lifetime equivalent consumption of fewer than 100 cigarettes. The definition of a radically treated patient was one who underwent an oncologic surgical procedure with removal of all macroscopic tumor, and intent to cure. Ethics approval was obtained from the Human Research and Ethics Committees at St Vincent’s Hospital and Peter MacCallum Cancer Centre, Melbourne.
Histologic Evaluation
The location, number, and size of tumors were obtained from the pathology reports. One pathologist, blinded to patient outcome, reviewed all hematoxylin and eosin (H&E)-stained slides. All cases were histologically classified according to the 2004 WHO Classification,28 apart from all adenocarcinomas, which were classified according to the new International Association for the Study of Lung Cancer (IASLC)/American Thoracic Society (ATS)/European Respiratory Society (ERS) international multidisciplinary lung adenocarcinoma classification.30 All squamous cell carcinomas were further subtyped into the variants according to the 2004 WHO Classification.28 In combined tumors with an admixture of different tumor subtypes, each subtype was estimated in 10% increments. Visceral pleural invasion was assessed in all cases, using guidelines of the 7th edition TNM classification.29, 31 All cases were examined for vascular, lymphatic channel, and perineural invasion, and, if included, hilar and mediastinal lymph nodes were examined to assess nodal disease status.
Immunohistochemical Evaluation
In accordance with recent IASLC/ATS/ERS recommendations30 regarding immunotyping of morphologically undifferentiated tumor in small biopsies and cytology specimens, which is further supported by recent work suggesting that resected large cell carcinomas harbor driver mutations in line with this immunotyping,32 all cases classified as large cell carcinoma underwent immunostaining with a panel of antibodies: TTF1 (SPT24, NovoCastra, Newcastle Upon Tyne, UK, 1:100 dilution); TP63 (4A4, DakoCytomation, Glostrup, Denmark, 1:300 dilution); AE1/AE3 (cocktail of AE1 and AE3, ratio of 20:1, NovoCastra, 1:80 dilution); cytokeratin (CK) 5 (XM26, NovoCastra, 1:50 dilution); CK7 (OV-TL 12/30, DakoCytomation, 1:200 dilution); epithelial membrane antigen (E29, DakoCytomation, 1:100 dilution); CD56 (Ventana Automatic, Tucson, AZ, USA); chromogranin A (DakoCytomation, 1:600 dilution); and synaptophysin (Ventana Automatic). In line with Rekhtman et al,32 TP63- and TTF1-negative large cell carcinomas and pleomorphic carcinomas were designated as ‘marker-null’ to indicate their lack of squamous and glandular morphology and differentiation. All cases classified as large cell neuroendocrine carcinoma, small cell carcinoma, typical and atypical carcinoid, pleomorphic carcinoma, and adenosquamous carcinoma underwent immunostaining with the same antibody panel used in large cell carcinomas. Most squamous cell carcinomas and adenocarcinomas also underwent TTF1, TP63, CK7, and CK5 immunostaining. Immunostaining was performed as per the manufacturer’s protocols on a Ventana Benchmark XT automated immunostainer (Ventana Medical Systems, Tucson, AZ, USA).
Tissue Microarrays
Formalin-fixed paraffin-embedded tumor blocks were obtained for all 388 tumors. A pathologist identified and circled three areas containing the largest proportion of the predominant tumor subtype on a corresponding H&E-stained slide and, if present, one area each of any lesser tumor subtypes and an area of normal tissue, with each tumor represented by at least four cores. Representative 1-mm tumor cores were punched from these areas for creation of tissue microarray blocks, as previously described.15
FGFR1 FISH
FGFR1 FISH assays were performed on four-micron unstained sections cut from each formalin-fixed paraffin-embedded block from the tissue microarray or from whole sections with the FISH assay conditions with FGFR1/CEN8 probe mix (Abnova Corporation, Taipei City, Taiwan), as previously described.15
A scientist experienced with FISH and a pathologist separately scored the FGFR1 FISH slides on a Leica DM60008 upright fluorescence microscope. For each slide, individual tumor cores or previously defined areas of tumor on whole sections were indentified at low ( × 10) magnification using the DAPI filter. In line with the standardized reading and evaluation strategy proposed by Schildhaus et al,26 tumors were only assessed if normal tissues including blood vessels, fibroblasts, or nontumor lung contained one or two distinct signals of each color. Tumors were scanned with an × 100 oil-immersion objective for amplification hot spots, which were chosen for assessment. If hot spots were not identified, random areas were chosen. A minimum of 60 tumor cells, comprising 10–15 contiguous cells from at least six fields, were photographed and scored for the number of FGFR1 (red) and chromosome 8 (green) signals. The FGFR1/CEN8 ratio, the number and percentage of cells with ≥5 and ≥15 FGFR1 signals, the average number of FGFR1 copy number/cell, and the number of CEN8 signals were calculated.
We used the terminology proposed by Schildhaus et al26 of high-level FGFR1 amplification for cases with an FGFR1/CEN8 ratio ≥2, or tumor cell percentage with ≥15 FGFR1 signals ≥10%, or average number of FGFR1 signals/tumor cell nucleus ≥6, and low-level FGFR1 amplification as tumor cell percentage with ≥5 FGFR1 signals ≥50%. This latter category of low-level FGFR1 amplification has been termed gene copy number gain by others.23 Owing to the absence of an internationally accepted definition of CEN8 polysomy, we arbitrarily defined it as ≥4 CEN8 signals in ≥40% of tumor cell nuclei.
FGFR1 SISH
FGFR1 SISH assays were performed on 4-micron unstained sections cut from each formalin-fixed paraffin-embedded block from the tissue microarray or whole sections. FGFR1 SISH was performed on an automated Ventana BenchMark XT platform (Ventana/Roche), with testing performed strictly according to the manufacturer’s protocols and by using recommended accessory reagents. Tissue sections were deparaffinized, denatured, and pretreated with cell conditioner no. 2 (Ventana) in four cycles of 8 min at 90 °C and incubated for 20 min with ISH Protease 3 (Ventana). The FGFR1 dinitrophenyl (DNP) probe combined with chromosome 8 digoxigenin (DIG) probe was added, and slides were incubated for 8 min and then denatured and incubated for 20 min at 80 °C. Slides were hybridized for 6 h at 44 °C and washed stringently three times at 72 °C for 8 min. Silver detection was achieved by incubation with ultraView silver ISH DNP rabbit anti-DNP antibody (Ventana) for 20 min, then by incubation with silver ISH DNP HRP for 16 min, and finally by incubation for cycles of 4 min with silver ISH DNP chromogens A and B and for 16 min with chromogen C. Red detection was achieved by incubation with Red ISH DIG mouse antibody for 20 min, followed by incubation with RED ISH DIG AP for 24 min. Slides were then incubated with Red ISH DIG pH enhancer for 8 min, Red ISH DIG naphthol for 4 min, and Red ISH DIG fast red for 4 and then 8 min. Slides were counterstained with Hematoxylin II (Ventana), incubated for 8 min, then post-counterstained with Bluing Reagent (Ventana), and incubated for 4 min. After the washing steps, the slides were mounted with a nonoxidative mounting medium (Pertex, Medite GmbH, Burgdorf, Germany).
One pathologist blinded to the location of the FGFR1-amplified tumors in the tissue microarray scored the FGFR1 SISH. Tumors were scanned for hot spots. If hot spots were not identified, random areas for assessment were chosen. A minimum of 60 tumor cells comprising 10–15 contiguous cells from six fields were assessed for the number of FGFR1 (black) and chromosome 8 (red) signals. The FGFR1/CEN8 ratio, the number of cells with ≥5 and ≥15 FGFR1 signals, and the average number of FGFR1 copy number/cell were calculated.
Statistical Evaluation
Comparisons of odds or proportions were made using the χ2 test. CIs were set at 95%. Analysis of variance was used to detect any correlation between FGFR1 amplification status and other variables. Survival was calculated from the date of pathological diagnosis until death, with cases lost to follow-up censored at date of last documented contact. Only patients who were surgically radically treated with curative intent were included in survival analyses, whereas those who underwent a biopsy or limited surgical resection for diagnosis without curative intent were not included. Differences in survival curves were determined by the log rank test. Statistical significance was set at P<0.05 for all analyses. Potential survival differences by covariates were calculated using the Cox regression. Survival plots were created using RStudio version 0.97.551.
Results
Patients and Tumors
The 352 evaluable patients comprised 63% (n=222) men, 20% current smokers, 63% former smokers, and 10% never smokers, with a median age of 69 years (range, 19–87 years) (Table 1). There were 46% stage I, 28% stage II, 21% stage III, and 4% stage IV patients. Smoking status and stage were unknown in 7% and 1% of patients, respectively. The 352 patients had 368 tumors able to be assessed by FISH, 11 patients with two tumors tested, and 1 patient with three metachronous tumors tested. The majority of patients (97%; n=342) underwent surgical resection, 92% with concomitant hilar and mediastinal lymph node dissection (n=313) (Table 1), whereas 11 patients had biopsies only, 10 from the lung primary and 1 from a left renal metastasis. The status of visceral pleural invasion, lymphatic channel, vascular space, and perineural invasion for the 357 resected tumors are shown in Table 2.
The 368 tumors able to be assessed by FISH comprised 225 tumors (61%) with either a morphologic component or immunoprofile of squamous cell carcinoma, including 178 pure squamous cell carcinomas; 23 large cell carcinomas with squamous immunoprofile;30, 33 18 adenosquamous carcinomas; and 6 pleomorphic carcinomas, 5 with ≥10% morphologic squamous component and 1 with ≥10% of large cell carcinoma with squamous immunoprofile, all admixed with ≥10% malignant spindle or giant cell carcinoma component (Table 2). The 143 nonsquamous tumors (39%) comprised 99 adenocarcinomas; 18 large cell carcinomas comprising eight ‘marker-null’ large cell carcinomas,30, 32 7 with an immunoprofile of lung adenocarcinoma,30, 33 and 3 large cell neuroendocrine carcinomas; 12 pleomorphic carcinomas, all without squamous component or immunoprofile; 5 small cell carcinomas; 5 carcinoid tumors; and 4 combined tumors with various admixtures of adenocarcinoma, small cell carcinoma, and large cell neuroendocrine carcinoma (Table 2).
FGFR1 FISH Amplification
FGFR1 FISH was performed in 406 tumors, and was assessable in 368 (90.6%) with 50 tumors (13.6%), from 49 patients, positive for FGFR1 amplification (Table 2 and Supplementary Table 1). The 50 FGFR1-amplified tumors comprised 40 pure squamous cell carcinomas; 5 large cell carcinomas with squamous immunoprofile; 2 adenosquamous carcinomas; 1 pleomorphic carcinoma with 55% squamous cell carcinoma, 30% giant cell/spindle cell carcinoma, and 15% adenocarcinoma; one ‘marker-null’ pleomorphic carcinoma comprising 80% giant cell carcinoma and 20% large cell carcinoma; and one ‘marker-null’ large cell carcinoma.
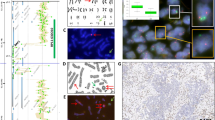
Of 225 assessable tumors with either squamous morphology or immunoprofile, there were 48 tumors (21.3%) with FGFR1 amplification (Table 2). In addition, of 143 assessable nonsquamous tumors, there were 2 tumors (1.4%) with FGFR1 amplification (Table 2). The relationship between FGFR1 amplification status and the presence or absence of squamous morphology or immunoprofile was statistically significant (P<0.0001). All other tumors tested by FGFR1 FISH were negative (Figure 1a). Of 50 FGFR1-amplified tumors, the frequency of high-level amplification was 82% (n=41), and low-level amplification was 18% (n=9) (Supplementary Table 1) (Figures 1b–d). In most FGFR1-amplified cases, we observed focal rather than homogeneous FGFR1 amplification, with occasional colocalized clusters of increased FGFR1 and CEN8 signals seen, with most not qualifying for our definition of CEN8 polysomy.
Representative photomicrographs of FGFR1 (red) and chromosome 8 (green) signal patterns. (a) A homogeneously nonamplified tumor with 1–2 FGFR1 signals per nucleus. (b) A tumor cell nucleus with high-level amplification with ≥15 FGFR1 signals scattered throughout the nucleus. (c) Homogeneous high-level amplification in a tumor with ≥6 FGFR1 signals per nucleus. (d) Low-level amplification in a tumor with several microclusters consisting of ≥5 FGFR1 signals.
Among 225 assessable tumors with either squamous morphology or immunoprofile, the average FGFR1 copy number/nucleus ranged from 1.1 to 12.3 (mean: 4.1). The mean FGFR1/CEN8 ratio was 1.4 (range: 0.5–6.1), and the mean percentages of tumor cells with ≥5 and ≥15 FGFR1 copies were 23% and 3.8%, respectively. Of 225 assessable tumors with either squamous morphology or immunoprofile, the frequency of high-level amplification was 17.3% (n=39) and low-level amplification was 4% (n=9).
Among the 50 FGFR1-amplified tumors, FGFR1/CEN8 ≥2 was the most frequent criterion of high-level amplification seen in 40 of 41 tumors, followed by ≥15 FGFR1 signals in ≥10% tumor cell nuclei in 22 of 41 tumors, and ≥6 FGFR1 signals/tumor cell nucleus in 20 of 41 tumors (Supplementary Table 1). One high-level amplification tumor with an FGFR1/CEN8 ratio <2 showed ≥15 FGFR1 in 44% of tumor nuclei and CEN8 polysomy (Supplementary Table 1). There were four other FGFR1-amplified tumors with CEN8 polysomy, all with ≥5 FGFR1 signals in ≥50% tumor nuclei but FGFR1/CEN8 ratio <2, thus qualifying for low-level amplification only. Otherwise, CEN8 polysomy was rarely seen in nonamplified tumors.
Of interest, the patient with three metachronous tumors had two FGFR1-amplified tumors that were both basaloid variant squamous cell carcinomas, whereas the third tumor was a poorly differentiated squamous cell carcinoma without basaloid morphology that was FISH negative. Another patient with an FGFR1-amplified moderately differentiated squamous cell carcinoma had an adenosquamous carcinoma 4 years previously that was FISH negative.
FGFR1 SISH Amplification
FGFR1 SISH was performed in 385 tumors, with 90.1% of tumors (n=347) assessable. FGFR1 amplification as detected by SISH was present in 46 of 50 FGFR1 FISH-amplified tumors that were tested (Figure 2). Four of 50 FGFR1-amplified tumors were unable to be tested for FGFR1 SISH, as we did not have sufficient tumor tissue for testing. All other tumors tested, which were negative for FGFR1 amplification by FISH, were negative for FGFR1 amplification by SISH.
Correlation of FGFR1 Amplification and Clinicopathologic Parameters
Postoperative 30-day mortality was 3.0% (10 of 332) and median follow-up was 30.2 months (range, 1–139 months). Overall survival for surgically radically treated patients with FGFR1-amplified tumors was similar to those treated radically surgically with FGFR1-negative tumors, when comparing patients with pure squamous cell carcinoma only (n=172; log rank P=0.50, HR=1.17, 95% CI=0.73–1.89); when comparing patients whose tumors had either any squamous morphology or immunoprofile (n=213; log rank P=0.97, HR=1.01, 95% CI=0.65–1.58) (Figure 3); when comparing all patients with non-small cell lung carcinoma (n=330; log rank P=0.66, HR=1.09, 95% CI=0.72–1.66); and when comparing all patients with lung malignancy, excluding five patients with surgically radically treated carcinoid tumors (n=341; log rank P=0.70, HR=1.08, 95% CI=0.71–1.63). Disease-free survival for surgically radically treated patients whose tumors had any squamous morphology or immunoprofile was also no different between those with FGFR1-amplified and nonamplified tumors (n=213; log rank P=0.88, HR 1.04, 95% CI 0.67–1.60). There was also no survival difference between surgically radically treated patients when stratified according to stage and FGFR1 amplification status (Table 3; Figure 4). The following variables were analyzed for the survival function of patients with tumors with either squamous morphology or immunoprofile: FGFR1 amplification, smoking history, age, sex, tumor size, node status, and stage (7th revision TNM). Significant variables included age (P=0.029), tumor size (P=0.021), stage (P<0.0001), and node status (P<0.0001). When these variables were entered into multivariate analysis, only age (P=0.039) and node status (P=0.049) remained significant. There was a statistically significant relationship between sex and FGFR1 amplification status (P=0.025) among all patients with non-small cell lung carcinoma, such that more men had FGFR1-amplified tumors than women (Table 1), although significance was lost if only patients with tumors with either squamous morphology or immunoprofile were examined (P=0.20). There was a statistically significant relationship between smoking and FGFR1 amplification status (P=0.040) among all patients with non-small cell lung carcinoma who ever smoked, but not between current smokers (P=0.106), although significance was lost if only patients with tumors with either squamous morphology or immunoprofile were examined (P=0.137). No significant relationship was detected between FGFR1 amplification status and visceral pleural invasion status (P=0.48), lymphatic channel invasion status (P=0.65), vascular space invasion status (P=0.26), or perineural invasion status (P=0.47).
Discussion
In this study, we confirm that FGFR1 amplification as detected by FISH is a frequent abnormality in squamous cell carcinoma and tumors with focal squamous morphology or immunoprofile in a large cohort of Australian lung cancer patients (48 of 225; 21.3%). FGFR1 amplification was also identified in 2 of 143 nonsquamous cases (1.4%), both ‘marker-null’ tumors without squamous or glandular morphology or immunoprofile. This difference was statistically significant (P<0.0001). Furthermore, when we compared overall and disease-free survival between surgically radically treated patients with pure squamous cell carcinoma, those with tumors with either focal squamous morphology or immunoprofile, those with non-small cell lung carcinoma, and those with any lung malignancy, with and without FGFR1 amplification, we found no statistical difference. We also found no significant difference in overall survival in surgically radically treated patients stratified by stage and FGFR1 amplification status. In addition, we validated SISH as a method to detect FGFR1 amplification, demonstrating a 1:1 correlation with FISH.
Our results are consistent with the two initial studies reporting FGFR1 amplification preferentially in squamous cell carcinoma with few amplified adenocarcinomas detected (22% squamous cell carcinoma and 1% nonsquamous tumors by FISH from Weiss et al;9 21% squamous cell carcinoma and 3.4% adenocarcinoma (P<0.001) by SNP arrays from Dutt et al10). The identification of this potentially actionable molecular target in a significant proportion of non-small cell lung carcinomas highlights the importance of defining FGFR1 amplification and the detection method used. A validated definition of FGFR1 amplification is an important step in the design of recently commenced phase I clinical trials evaluating FGFR1 inhibitors. As with other laboratory tests, the method used to identify FGFR1 amplification must provide a result within a clinically meaningful time frame, be affordable, robust, and valid in small biopsy samples.
Because we performed detailed morphologic classification on all and immunotyping on most tumors in our data set, we found that not only does FGFR1 amplification occur most commonly in squamous cell carcinoma but it also occurs in tumors with focal squamous morphology or immunoprofile. Schildhaus et al26 reported one FGFR1-amplified adenosquamous carcinoma in their cohort but unfortunately no other group provided immunotyping details of large cell carcinomas tested, apart from one study that reported TTF1 results (but not TP63) in nearly all large cell carcinomas.23 All but one FGFR1-amplified large cell carcinoma in our cohort were TP63-positive and TTF1-negative, whereas no TTF1-positive large cell carcinomas tested were amplified. In addition, we detected FGFR1 amplification in two ‘marker-null’ tumors, a large cell carcinoma, and a pleomorphic carcinoma, both lacking squamous or glandular morphology or immunoprofile. These findings have great importance for the molecular taxonomy of large cell carcinoma in resected specimens, as they confirm that large cell carcinomas with squamous immunoprofile have a genomic aberration in common with squamous cell carcinoma, and suggest that such tumors should be considered for classification with squamous cell carcinoma in future lung tumor classifications. In addition, ‘marker-null’ tumors including large cell carcinomas and pleomorphic carcinomas can be FGFR1-amplified too, suggesting common squamous origins. Our findings also support the significance of recent work that has emphasized the relevance of immunotyping resected large cell carcinoma by demonstrating that the driver mutation profiles present were in line with the immunotyping results.32 Furthermore, we propose that not only should all tumors with squamous morphology and/or immunoprofile be tested for FGFR1 amplification but so should all ‘marker-null’ large cell carcinomas and pleomorphic carcinomas.
FISH has been the most frequently used method to detect FGFR1 amplification status,9, 10, 21, 22, 25, 26, 27 although alternative methods such as real-time polymerase chain reaction24 and SISH23 have also been used. SISH has several advantages over FISH including automation, use of bright-field microscopes in contrast to expensive FISH microscopes, signal permanence, and easier histologic comparison. If our finding of 1:1 correlation of FGFR1 amplification status between FISH and SISH is subsequently validated, it indicates that SISH could be used to determine FGFR1 amplification in the future, either as a screening tool to select cases for FISH testing or as a stand-alone detection method in centers where the more expensive FISH technique is not available.
The definition of FGFR1 amplification varies in different studies, some using the same method of detection. Many studies9, 22, 27 using FISH to detect FGFR1 amplification used the following definitions: >9 signals/cell for high-level amplification, >2 but <9 signals/cell for low-level amplification, both of which were amended by one group such that those copy numbers must be in excess of the CEN8 signals in ≥20% of nuclei,27 and <2 signals/cell as not amplified. Another study using FISH introduced a definition of FGFR1 gene to copy number ratio of ≥2.2,21 whereas another used a definition of ≥4 signals/cell for FGFR1 ‘FISH positivity’, reporting no examples of higher level amplification, which was defined as ≥9 signals/cell.25 We chose to investigate the standardized reading and evaluation strategy and evaluation criteria for FGFR1 FISH proposed by Schildhaus et al,26 as we have performed FGFR1 FISH for clinical reasons in our department for some months and had noticed some of the FGFR1 amplification patterns detailed in their report. Moreover, as Schildhaus et al point out in their study, the initial validated data suggested that cancer cell lines with focal high-level amplification respond better to FGFR1 inhibitors9, 10 than those with low-level amplification, and thus it may be that detailed definitions of high-level amplification, as proposed by Schildhaus et al26 and validated here by us, are more relevant than low-level amplification, which is examined in more detail by other groups.23, 25 Ultimately the definition of FGFR1 amplification will need to be determined according to treatment response, and further studies validating the proposals by Schildhaus et al are needed.
No significant relationships between FGFR1 amplification status and overall and disease-free survival were detected among surgically radically treated patients in our cohort when clustered into four different groups, including those with pure squamous cell carcinomas only, those with tumors with any squamous morphology or immunoprofile, those with non-small cell lung carcinomas, and those with any lung malignancy. We divided our patient cohort into these groups as other studies investigating FGFR1 amplification in non-small cell lung carcinomas include patients with squamous cell carcinoma only21, 22, 27 and patients with squamous cell carcinoma, adenocarcinoma, and large cell carcinoma.23, 24, 25, 26 In addition, we found no significant difference in overall survival in surgically radically treated patients stratified by stage and FGFR1 amplification status. Considering statistically significant results only, most studies9, 21, 24, 25, 26 reported a lack of a significant relationship between survival and FGFR1 amplification status, in line with our findings. However, two studies did detect an effect of FGFR1 amplification on survival with Kim et al22 reporting significantly greater risk of recurrence and death for patients with FGFR1-amplified squamous cell carcinoma, whereas Tran et al23 reported a nonsignificant direction of effect toward longer overall survival for patients with FGFR1-amplified tumors in univariate analysis (P=0.14), which became significant on multivariate analysis (HR 0.6, P=0.02). The difficulty with interpreting the latter two results is that each study used different definitions for FGFR1 amplification and different detection methods. More studies using the definitions proposed by Schildhaus et al are needed to resolve the question of a significant relationship between survival and FGFR1 amplification status.
In conclusion, FGFR1 amplification as detected by FISH is a frequent targetable abnormality in squamous cell carcinomas and in tumors with any squamous morphology or immunoprofile, but may also occur in ‘marker-null’ large cell carcinomas and pleomorphic carcinomas. No survival differences according to FGFR1 amplification status were apparent in our cohort of surgically radically treated patients. We have validated SISH as a method to detect FGFR1 amplification, demonstrating a 1:1 correlation with FISH, indicating that SISH may be an alternative method to detect FGFR1 amplification.
References
Australian Institute of Health and Welfare & Australasian Association of Cancer Registries 2012. Cancer in Australia: An Overview . Cancer Series No. 74. Cat. No. CAN 70. AIHW: Canberra, 2012.
Mok TS, Wu YL, Thongprasert S et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947–957.
Maemondo M, Inoue A, Kobayashi K et al. North-East Japan Study Group. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380–2388.
Mitsudomi T, Morita S, Yatabe Y et al. West Japan Oncology Group. Gefinitib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomized phase 3 trial. Lancet Oncol 2010;11:121–128.
Rosell R, Carcereny E, Gervais R et al. Spanish Lung Cancer Group in collaboration with Groupe Francais de Pneumo-Cancerologie and Associazone Italiana Oncologia Toracica. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomized phase 3 trial. Lancet Oncol 2012;13:239–246.
Han JY, Park K, Kim SW et al. First-SIGNAL: first-line single-agent Iressa Versus Gemcitabine and Cisplatin Trial in never-smokers with adenocarcinoma of the lung. J Clin Oncol 2012;30:1122–1128.
Zhou C, Wu YL, Chen G et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomized phase 3 study. Lancet Oncol 2011;12:735–742.
Kwak EL, Bang YJ, Camidge DR et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693–1703.
Weiss J, Sos ML, Seidel D et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;15:62ra93.
Dutt A, Ramos AH, Hammerman PS et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS ONE 2011;6:e20351.
Bass AJ, Watanabe H, Mermel CH et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet 2009;41:1238–1242.
Itoh N, Ornitz DM . Fibroblast growth factors: from molecular evolution to roles in development, metabolism and disease. J Biochem 2010;149:121–130.
Turner N, Grose R . Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 2010;10:116–129.
Brooks AN, Kilgour E, Smith PD . Molecular pathways: Fibroblast growth factor signalling – a new therapeutic opportunity in cancer. Clin Cancer Res 2012;18:1855–1862.
Young RJ, Lim AM, Angel C et al. Frequency of fibroblast growth factor receptor 1 gene amplification in oral tongue squamous cell carcinomas and associations with clinical features and patient outcome. Oral Oncol 2013;49:576–581.
Ishizuka T, Tanabe C, Sakamoto H et al. Gene amplification profiling of esophageal squamous cell carcinomas by DNA array CGH. Biochem Biophys Res Commun 2002;296:152–155.
Gorringe KL, Jacobs S, Thompson ER et al. High-resolution single nucleotide polymorphism array analysis of epithelial ovarian cancer reveals numerous microdeletions and amplifications. Clin Cancer Res 2007;13:4731–4739.
Simon R, Richter J, Wagner U et al. High-throughput tissue microarray analysis of 3p25 (RAF1) and 8p12 (FGFR1) copy number alterations in urinary bladder cancer. Cancer Res 2001;61:4514–4519.
Edwards J, Krishna NS, Witton CJ et al. Gene amplifications associated with the development of hormone-resistant prostate cancer. Clin Cancer Res 2003;9:5271–5281.
Missiaglia E, Selfe J, Hamdi M et al. Genomic imbalances in rhabdomyosarcoma cell lines affect expression of genes frequently altered in primary tumors: an approach to identify candidate genes involved in tumor development. Genes Chromosomes Cancer 2009;48:455–467.
Heist RS, Mino-Kenudson M, Sequist LV et al. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol 2012;7:1775–1780.
Kim HR, Kim DJ, Kang DR et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol 2013;31:73–77.
Tran TN, Selinger CI, Kohonen-Corish MR et al. Fibroblast growth factor receptor 1 (FGFR1) copy number is an independent prognostic factor in non-small cell lung cancer. Lung Cancer 2013;81:462–467.
Sasaki H, Shitara M, Yokota K et al. Increased FGFR1 copy number in lung squamous cell carcinomas. Mol Med Rep 2012;5:725–728.
Kohler LH, Mireskandari M, Knosel T et al. FGFR1 expression and gene copy numbers in human lung cancer. Virchows Arch 2012;461:49–57.
Schildhaus H-U, Heukamp LC, Merkelbach-Bruse S et al. Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol 2012;25:1473–1480.
Göke F, Franzen A, Menon R et al. Rationale for treatment of metastatic squamous cell carcinoma of the lung using fibroblast growth factor receptor inhibitors. Chest 2012;142:1020–1026.
Travis WD, Brambilla E . Chapter 1. Tumours of the lung In: Travis WD, Brambilla E, Muller-Hermelink HK, et al (eds). Pathology and Genetics: Tumours of the Lung, Pleura, Thymus and HeartWorld Health Organisation Classification of Tumours. IARC Press: Lyon, 2004, pp 26–62.
Goldstraw P et al. Chapter 5. Site-specific explanatory notes for lung and pleural tumours In: Goldstraw P, (ed) International Association for the Study of Lung Cancer Staging Manual in Thoracic Oncology. Editorial Rx Press: Orange Park, FL, 2009, pp 67–80.
Travis WD, Brambilla E, Noguchi M et al. The new IASLC/ATS/ERS international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol 2011;6:244–285.
Travis WD, Brambilla E, Rami-Porta R et al. Visceral pleural invasion: pathologic criteria and use of elastic stains: proposal for the 7th edition of the TNM classification for lung cancer. J Thorac Oncol 2008;3:1384–1390.
Rekhtman N, Tafe LJ, Chaft JE et al. Distinct profile of driver mutations and clinical features in immunomarker-defined subsets of pulmonary large-cell carcinoma. Mod Pathol 2013;26:511–522.
Rekhtman N, Ang DC, Sima CS et al. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod Pathol 2011;24:1348–1359.
Acknowledgements
This study was supported by a financial contribution from Collingwood Rotary Members. GMW was supported by National Health and Medical Research Council Scholarship GNT 1038699 and University of Melbourne Medical Postgraduate Committee Gordon-Taylor Scholarship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Russell, P., Yu, Y., Young, R. et al. Prevalence, morphology, and natural history of FGFR1-amplified lung cancer, including squamous cell carcinoma, detected by FISH and SISH. Mod Pathol 27, 1621–1631 (2014). https://doi.org/10.1038/modpathol.2014.71
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2014.71