Abstract
Glomangiopericytoma (sinonasal-type hemangiopericytoma) is a rare mesenchymal neoplasm with myoid phenotype (smooth muscle actin-positive), which distinguishes this tumor from soft tissue hemangiopericytoma/solitary fibrous tumor. Molecular genetic changes underlying the pathogenesis of glomangiopericytoma are not known. In this study, 13 well-characterized glomangiopericytomas were immunohistochemically evaluated for β-catenin expression. All analyzed tumors showed strong expression and nuclear accumulation of β-catenin. Following this observation, β-catenin glycogen serine kinase-3 beta phosphorylation region, encoded by exon 3, was PCR amplified in all cases and evaluated for mutations using Sanger sequencing. Heterozygous mutations were identified in 12 of 13 tumors. All mutations consisted of single-nucleotide substitutions: three in codon 32 (c.94G>C (n=2) and c.95A>T), four in codon 33 (two each c.98C>G and c.98C>T), two in codon 37 (c.109T>G), one in codon 41 (c.121A>G), and two in codon 45 (c.133T>C). At the protein level, these substitutions would lead to p.D32H, p.D32V, p.S33C, p.S33F, p.S37A, p.T41A, and p.S45L mutations, respectively. Previously, similar mutations have been reported in different types of cancers and shown to trigger activation of β-catenin signaling. All analyzed glomangiopericytomas showed prominent nuclear expression of cyclin D1, as previously shown for tumors with nuclear expression of β-catenin as a sign of oncogenic activation. These results demonstrate that mutational activation of β-catenin and associated cyclin D1 overexpression may be central events in the pathogenesis of glomangiopericytoma. In additon, nuclear accumulation of β-catenin is a diagnostic marker for glomangiopericytoma.
Similar content being viewed by others
Main
Glomangiopericytoma (sinonasal-type hemangiopericytoma) is a rare mesenchymal neoplasm of the nasal cavity and sinuses of low biologic potential.1, 2, 3, 4 It shows a myoid phenotype with consistent expression of smooth muscle actin.3
β-Catenin, a cadherin-associated membrane protein, participates in the regulation of cell-to-cell adhesion and in some circumstances, also gene transcription as a nuclear protein, a terminal component of the canonical Wnt-signaling pathway. Aberrant expression of β-catenin encoded by CTNNB1 gene is a well-known event in tumorigenesis and tumor progression.5, 6
Among soft tissue tumors, somatic mutations in CTNNB1 are well known in the pathogenesis of sporadic desmoid-type fibromatosis.7, 8 These mutations cluster in CTNNB1 glycogen serine kinase-3 beta (GSK3β) phosphorylation region and constitutionally activate β-catenin signal by upholding cellular β-catenin levels. This happens via interference of ubiquitin-mediated proteolytic degradation by elimination of the phosphorylation sites necessary for ubiquitin action.7, 8 Accumulation of β-catenin in turn results in nuclear translocation, which is a telltale sign of a mutation, typically seen in sporadic desmoid-type fibromatosis and other tumors driven by CTNNB1 mutations and useful in immunohistochemical diagnosis.9 Similar nuclear accumulation of β-catenin with CTNNB1 mutations also occurs in nasopharyngeal angiofibroma.10
On the basis of an initial observation of β-catenin expression in one case of glomangiopericytoma, we examined immunohistochemically β-catenin expression and searched for DNA mutations in CTNNB1 in a series of glomangiopericytomas.
Materials and methods
Thirteen tumors diagnosed as glomangiopericytoma (sinonasal-type hemangiopericytoma) were selected for this study. A majority of these tumors were previously characterized clinically and immunophenotypically.3
Immunohistochemistry
Immunohistochemistry for β-catenin, cyclin D1, CD34, desmin, smooth muscle actin, S100 protein, STAT6, and vimentin was performed using Leica Bond-Max automatic immunostainer (Bannockburn, IL). Appropriate positive and negative controls were included for all immunohstochemical reactions. Antibodies and conditions are listed in Table 1. Immunohistochemical staining was scored as negative or positive, and percentage of positive cells was estimated.
DNA Extraction
Formalin-fixed paraffin-embedded (FFPE) tumor tissue was available for DNA extraction in all cases. One to ten 5 μm thick sections (depending on the sample size) were deparaffinized with xylene, washed twice in ethanol, lyophilized, and incubated with 10 μg/μl proteinase K (Roche Diagnostics, Indianapolis, IN) in Hirt-Buffer at 55 °C for at least 24 h. DNA was recovered using the Maxwell®16 robotic system and DNA IQ™ Casework Pro Kit for Maxwell® 16 following the manufacturer’s protocol (Promega, Madison, WI).
PCR Amplification for Sanger Sequencing
Mutational ‘hot spot’ in CTNNB1, a GSK3β phosphorylation region encoded by exon 3 was PCR amplified using AmpliTaq Gold® DNA polymerase (Applied Biosystems, Roche, Branchburg, NJ) and two primers, CTNNB1.3 forward 5′-GACAGAAAAGCGGCTGTTAG-3′ and CTNNB1.3 reverse 5′-ACATCCTCTTCCTCAGGATT-3′ following standard three-temperature PCR protocol with denaturing at 95 °C for 30 s, annealing at 48 °C for 45 s, and extension at 72 °C for 45 s. In all, 50 μl PCRs were evaluated on 2% agarose gels. PCR products were extracted and purified using the QIAquick Gel Extraction Kit (www.qiagen.com). Sanger sequencing of these products was performed with forward and reverse primers by MacrogenUSA (www.macrogenusa.com) Obtained sequences were analyzed and aligned with CTNNB1 reference sequence, NG_013302.1 (www.ncbi.nlm.nih.gov). All PCR and sequencing experiments were repeated at least two times.
Results
Demographic and Clinicopathologic Data
Tumors from 8 male and 5 female patients were included in this study. Both the mean and median age of the patients was 74 years. All tumors were polypoid masses involving the nasal cavities (7 in the right cavity, 3 in the left cavity, and 3 in unspecified nasal location). One tumor also involved the ethmoid sinus.
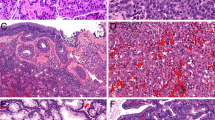
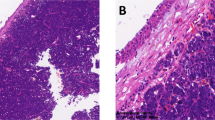
Histologically, these tumors consisted of epithelioid-to-spindled cells with a lightly eosinophilic cytoplasm and occasional peripheral clearing. Prominent blood vessels with gaping lumens were typically present, showing peritheliomatous hyalinization and sometimes pericellular clearing (Figure 1a–c). One tumor contained multinucleated giant cells (Figure 1d).
Immunohistochemistry Results
Strong nuclear β-catenin expression in virtually 100% of tumor cells was noted in all cases and this was typically accompanied by cytoplasmic staining (Figure 2). Similarly, cyclin D1 expression was seen in a great majority of tumor cells (70–100%) in all cases. Immunohistochemically, all cases were uniformly positive for vimentin. Smooth muscle actin was variably expressed in all but one case (30–100%). CD34 was variably expressed in all cases (5–100%, median 30%). All tumors were negative for desmin, S100 protein, and STAT6.
Molecular Genetics Findings
Mutations in the CTNNB1 gene encoding β-catenin were identified in 12 of the 13 analyzed tumors. Representative examples of Sanger sequencing are shown in Figure 3. All mutations consisted of single-nucleotide substitutions: three in codon 32 (c.94G>C (n=2) and c.95A>T), four in codon 33 (two each c.98C>G and c.98C>T), two in codon 37 (c.109T>G), one in codon 41 (c.121A>G), and two in codon 45 (c.133T>C). At the protein level, these substitutions would lead to p.D32H, p.D32V, p.S33C, p.S33F, p.S37A, p.T41A, and p.S45L mutations, respectively. Only wild-type (WT) CTNNB1 sequences were identified in one tumor. The results were reproducible in independent PCR experiments.
Discussion
Glomangiopericytoma (sinonasal hemangiopericytoma) is a rare mesenchymal tumor specific to sinonasal passages. It is diagnosed by characteristic histology showing epithelioid cells in a perivascular pattern with frequent perivascular hyalinization, and in most cases also by immunoreactivity for smooth muscle actin with variable positivity for CD34.1, 3
In this study we detected consistent, strong nuclear β-catenin expression in glomangiopericytoma and gain-of-function CTNNB1 mutations in nearly all cases studied. CTNNB1 mutations consisted of single-nucleotide substitutions affecting phosphorylation sites for GSK-3β, serine at codons 33, 37, and 45 and threonine at codon 41. However, in three cases neighboring aspartic acid at codon 32 was found to be affected. It has been proposed that mutations at the codons flanking GSK-3β phosphorylation sites may change the protein structure and, thereby affect the recognition by the ubiquitin-dependent proteolysis system responsible for degradation of β-catenin.11
In one glomangiopericytoma with strong nuclear β-catenin accumulation only WT CTNNB1 exon 3 sequence was identified. In this case, other genetic or epigenetic mechanisms leading to nuclear accumulation of β-catenin might be responsible. However, false negative results generated by PCR assay used in this study cannot be excluded. Various size deletions extending 5′ and 3′ from mutational ‘hot spot’ CTNNB1 GSK-3β region into intron 2 or intron 3 and exon 4, respectively, have been reported in other tumors.12, 13 Identification of such deletions would require well-preserved DNA and PCR assay designed to amplify ∼800 base pair (bp) DNA target. PCR assay used in this study was designed to amplify short targets (<125 bp) from severely degraded DNA from archival FFPE tissues. Thus, in a case of several hundred bp deletion priming sequences of this assay would be eliminated from mutant allele and only the WT allele would be amplified (pseudo wild type).
CTNNB1 mutations identical to those reported in this study have been previously identified in spectrum of epithelial, mesenchymal, and neuroectodermal tumors including, among others, hepatocellular carcinoma, desmoid-type fibromatosis and primitive neuroectodermal tumor (PNET). A complete list of references is available online from COSMIC, the catalogue of somatic mutations in cancer (http://cancer.sanger.ac.uk).
Several studies, including in vitro mouse models of hepatic carcinogenesis, have shown that abnormal nuclear expression of CTNNB1 can lead to cyclin D1 (CCND1) overexpression and thereby disregulate the cell cycle contributing to neoplastic transformation.14, 15, 16, 17, 18 In this study, all glomangiopericytomas showed nuclear expression of cyclin D1 indicating that CTNNB1 mutations and β-catenin overexpression in this tumor are also coupled with cell-cycle disregulation in this tumor.
The differential diagnosis for glomangiopericytoma especially includes solitary fibrous tumor and glomus tumor. In addition to histology1, 2, 3 and previously known immunohistochemical profile, β-catenin expression and mutation status can be a useful discriminator.
Solitary fibrous tumor is typically strongly and uniformly CD34 positive and SMA negative. Nuclear β-catenin expression has been reported in a subset of solitary fibrous tumors (SFT) from different locations, although mutation status has not been established.19, 20, 21, 22, 23 Additionally, SFT is characterized by NAB2-STAT6 gene fusion and overexpression of the fusion protein.24, 25 In this study, all glomangiopericytomas were immunohistochemically negative for STAT6 similar to the results recently reported in another study cohort, where all glomangiopericytomas were also negative for NAB2-STAT6 fusion gene transcripts.26 Therefore, STAT6 expression is also discriminatory between these entities.
Glomus tumor shares with glomangiopericytoma many histologic and immunohistochemical similarities, including the perivascular histologic pattern and SMA expression.3 However, MIR143-NOTCH gene fusion often seen in glomus tumor has not been found in glomangiopericytoma.27 In this study, further differences between glomangiopericytoma and glomus tumor included nuclear β-catenin expression and gain-of-function CTNNB1 mutations in glomangiopericytoma. Previous studies on glomus tumor, by contrast, showed a lack of β-catenin nuclear expression and CTNNB1 activating mutations.19, 28 Therefore, nuclear β-catenin expression and the presence of CTNNB1 mutations can be useful in distinguishing glomangiopericytoma and glomus tumor.
Fibroblastic neoplasms with nuclear β-catenin expression and CTNNB1 mutations include desmoid-type fibromatosis and nasopharyngeal angiofibroma. However, these tumors are histologically distinctly different in their spindle cell morphology and associated prominent collagenous matrix not present in glomangiopericytoma.9, 10
In summary, this study identifies oncogenic CTNNB1 mutations activating β-catenin signaling in glomangiopericytoma demonstrating that activation of Wnt-signaling pathway and subsequent overexpression of cyclin D1 have a central role in the molecular pathogenesis of this tumor. In addition, β-catenin nuclear expression and the presence of CTNNB1 mutations can be differential diagnostic markers for glomangiopericytoma.
References
Compagno J, Hyams VJ . Hemangiopericytoma-like intranasal tumors. A clinicopathologic study of 23 cases. Am J Clin Pathol 1976;66:672–683.
Eichhorn JH, Dickersin GR, Bhan AK et al. Sinonasal hemangiopericytoma. A reassessment with electron microscopy, immunohistochemistry, and long-term follow-up. Am J Surg Pathol 1990;14:856–866.
Thompson LD, Miettinen M, Wenig BM . Sinonasal-type hemangiopericytoma: a clinicopathologic and immunophenotypic analysis of 104 cases showing perivascular myoid differentiation. Am J Surg Pathol 2003;27:737–749.
Kuo FY, Lin HC, Eng HL et al. Sinonasal hemangiopericytoma-like tumor with true pericytic myoid differentiation: a clinicopathologic and immunohistochemical study of five cases. Head Neck 2005;27:124–129.
Reya T, Clevers H . Wnt signalling in stem cells and cancer. Nature 2005;434:843–850.
Klaus A, Birchmeier W . Wnt signalling and its impact on development and cancer. Nat Rev Cancer 2008;8:387–398.
Tejpar S, Nollet F, Li C et al. Predominance of beta-catenin mutations and beta-catenin dysregulation in sporadic aggressive fibromatosis (desmoid tumor). Oncogene 1999;18:6615–6620.
Huss S, Nehles J, Binot E et al. β-catenin (CTNNB1) mutations and clinicopathological features of mesenteric desmoid-type fibromatosis. Histopathology 2013;62:294–304.
Bhattacharya B, Dilworth HP, Iacobuzio-Donahue C et al. Nuclear beta-catenin expression distinguishes deep fibromatosis from other benign and malignant fibroblastic and myofibroblastic lesions. Am J Surg Pathol 2005;29:653–659.
Abraham SC, Montgomery EA, Giardiello FM et al. Frequent beta-catenin mutations in juvenile nasopharyngeal angiofibromas. Am J Pathol 2001;158:1073–1078.
Polakis P . The oncogenic activation of beta-catenin. Curr Opin Genet Dev 1999;9:15–21.
Miyoshi Y, Iwao K, Nagasawa Y et al. Activation of the beta-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res 1998;58:2524–2527.
Takayasu H, Horie H, Hiyama E et al. Frequent deletions and mutations of the beta-catenin gene are associated with overexpression of cyclin D1 and fibronectin and poorly differentiated histology in childhood hepatoblastoma. Clin Cancer Res 2001;7:901–908.
Tetsu O, McCormick F . Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999;398:422–426.
Saegusa M, Hashimura M, Kuwata T et al. Beta-catenin simultaneously induces activation of the p53-p21WAF1 pathway and overexpression of cyclin D1 during squamous differentiation of endometrial carcinoma cells. Am J Pathol 2004;164:1739–1749.
Zhang J, Gill AJ, Issacs JD et al. The Wnt/β-catenin pathway drives increased cyclin D1 levels in lymph node metastasis in papillary thyroid cancer. Hum Pathol 2012;43:1044–1050.
Anna CH, Iida M, Sills RC et al. Expression of potential beta-catenin targets, cyclin D1, c-Jun, c-Myc, E-cadherin, and EGFR in chemically induced hepatocellular neoplasms from B6C3F1 mice. Toxicol Appl Pharmacol 2003;190:135–145.
Gotoh J, Obata M, Yoshie M et al. Cyclin D1 over-expression correlates with β-catenin activation, but not with H-ras mutations, and phosphorylation of Akt, GSK3β and ERK1/2 in mouse hepatic carcinogenesis. Carcinogenesis 2003;24:435–442.
Ng TL, Gown AM, Barry TS et al. Nuclear beta-catenin in mesenchymal tumors. Mod Pathol 2005;18:68–74.
Carlson JW, Fletcher CD . Immunohistochemistry for beta-catenin in the differential diagnosis of spindle cell lesions: analysis of a series and review of the literature. Histopathology 2007;51:509–514.
Rakheja D, Molberg KH, Roberts CA et al. Immunohistochemical expression of beta-catenin in solitary fibrous tumors. Arch Pathol Lab Med 2005;129:776–779.
Yamaguchi U, Hasegawa T, Masuda T et al. Differential diagnosis of gastrointestinal stromal tumor and other spindle cell tumors in the gastrointestinal tract based on immunohistochemical analysis. Virchows Arch 2004;445:142–150.
Andino L, Cagle PT, Murer B et al. Pleuropulmonary desmoid tumors: immunohistochemical comparison with solitary fibrous tumors and assessment of beta-catenin and cyclin D1 expression. Arch Pathol Lab Med 2006;130:1503–1509.
Chmielecki J, Crago AM, Rosenberg M et al. Whole-exome sequencing identifies a recurrent NAB2-STAT6 fusion in solitary fibrous tumors. Nat Genet 2013;45:131–132.
Robinson DR, Wu YM, Kalyana-Sundaram S et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet 2013;45:180–185.
Agaimy A, Barthelmeß S, Geddert H et al. Phenotypic and molecular distinctness of sinonasal hemangiopericytoma compared to solitary fibrous tumor of the sinonasal tract. Histopathology 2014;65:667–673.
Mosquera JM, Sboner A, Zhang L et al. Novel MIR143-NOTCH fusions in benign and malignant glomus tumors. Genes Chromosomes Cancer 2013;52:1075–1087.
Chakrapani A, Warrick A, Nelson D et al. BRAF and KRAS mutations in sporadic glomus tumors. Am J Dermatopathol 2012;34:533–535.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Lasota, J., Felisiak-Golabek, A., Aly, F. et al. Nuclear expression and gain-of-function β-catenin mutation in glomangiopericytoma (sinonasal-type hemangiopericytoma): insight into pathogenesis and a diagnostic marker. Mod Pathol 28, 715–720 (2015). https://doi.org/10.1038/modpathol.2014.161
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2014.161
This article is cited by
-
CTNNB1 mutation-driven hybrid tumor: desmoid fibromatosis with an unusual associated epithelioid component arising in association with a neuromuscular choristoma
Virchows Archiv (2024)
-
Histopathologic Diagnosis of Sinonasal Tumors: Challenges and the Importance of Establishing the Correct Diagnosis
Current Otorhinolaryngology Reports (2023)
-
Top 10 Significant Spindled Head and Neck Lesions to Scrutinze
Head and Neck Pathology (2023)
-
Update from the 5th Edition of the World Health Organization Classification of Head and Neck Tumors: Nasal Cavity, Paranasal Sinuses and Skull Base
Head and Neck Pathology (2022)
-
Combined Neuroendocrine and Squamous Cell Carcinoma of the Sinonasal Tract: A Morphologic and Immunohistochemical Analysis and Review of Literature
Head and Neck Pathology (2022)