Abstract
The success of crizotinib in ALK-positive patients has elicited efforts to find new oncogenic fusions in lung cancer. These efforts have led to the discovery of novel oncogenic fusion genes such as ROS1 and RET. However, the molecular and clinicopathologic characteristics associated with RET or ROS1 fusion, compared with ALK fusion-positive lung cancer, remain unclear. We accordingly analyzed the clinicopathologic characteristics of RET- and ROS1-fusion-positive lung adenocarcinomas. We further performed immunohistochemistry and fluorescence in situ hybridization analysis (FISH) in 15 cases of RET and 9 cases of ROS1 fusion tumors by identified NanoString’s nCounter screening. RET fusion-positive patients were younger in age, never-smokers, and in early T stage; ROS1 fusion-positive patients had a higher number of never-smokers compared with patients with quintuple-negative (EGFR−/KRAS−/ALK−/ROS1−/RET−) lung adenocarcinoma. Histologically, RET and ROS1 fusion tumors share the solid signet-ring cell and mucinous cribriform pattern, as previously mentioned in the histology of ALK fusion tumors. Therefore, it can be presumed that fusion gene-associated lung adenocarcinomas share similar histologic features. In immunohistochemistry, the majority of 15 RET and 9 ROS1 fusion-positive cases showed positivity of more than moderate intensity and cytoplasmic staining for RET and ROS1 proteins, respectively. In FISH, the majority of RET and ROS1 rearrangement showed two signal patterns such as one fusion signal and two separated green and orange signals (1F1G1O) and an isolated 3′ green signal pattern (1F1G). Our study has provided not only characteristics of fusion gene-associated histologic features but also a proposal for a future screening strategy that will enable clinicians to select cases needed to be checked for ROS1 and RET rearrangements based on clinicohistologic features.
Similar content being viewed by others
Main
Recently, chromosomal rearrangements involving receptor tyrosine kinases (RTKs) have emerged as important oncogenic drivers of lung cancer. In 2007, Soda et al1 identified a subset of NSCLCs harboring chromosomal translocations involving the anaplastic lymphoma receptor tyrosine kinase (ALK). ALK rearrangements have since been identified in ∼3–5% of patients with lung cancer.2, 3 ALK-positive lung cancer has unique clinicopathologic features and shows dramatic clinical response to ALK inhibitors such as crizotinib.2, 3 The success of crizotinib in ALK-positive patients has elicited efforts to find new oncogenic fusions in lung cancer.4 These efforts have led to the discovery of novel oncogenic fusion gene such as ROS1 and RET.
ROS1 rearrangements in lung cancer were first identified in the non-small cell cancer cell line (HCC78 cell line) in 2007.5 ROS1 rearrangements have been identified in ∼1–2% of patients with lung cancer.6 ROS1 fusion-positive tumors define a distinct molecular subtype of lung cancer with unique clinicopathologic features, as well as ALK-positive lung cancer. ROS1 rearrangements were reported in patients with a younger age, no history of smoking, Asian ethnicity, advanced stage, and adenocarcinoma on histology.7 Unexpectedly, the HCC78 cell line was sensitive to treatment with crizotinib because of homology of the tyrosine kinase domain of ALK and ROS1.8, 9 Indeed, the use of crizotinib in ROS1-rearranged lung cancer has exhibited significant clinical activity. Recently, several clinical trials are ongoing on ROS1-positive patients worldwide.6
In 2012, Ju et al10 reported the first case of a 33-year-old, never-smoker lung adenocarcinoma patient harboring RET rearrangement. To date, several cancer genome sequencing studies have discovered RET fusions in ∼1–2% of lung cancer.10, 11, 12, 13 RET fusions were the potential therapeutic targets of multitargeted kinase inhibitors, vandetanib, sunitinib, and sorafenib.11, 12, 13 Importantly, RET rearrangement is mutually exclusive with aberrations in EGFR, KRAS, ALK, HER2, and BRAF in lung cancer.10, 11, 12, 13 Several studies recently reported that RET fusion-positive tumors represent distinct clinicopathologic features, as well as molecular subset.14, 15 However, the molecular and clinicopathologic characteristics associated with RET fusion compared with ALK or ROS1 fusion-positive tumors are still unclear. In particular, characteristic morphologic features have not been investigated, and signal patterns of fluorescence in situ hybridization analysis (FISH) analysis as an effective tool for the detection of RET fusions have rarely been described.
Here, we analyzed the clinicopathologic characteristics of RET- and ROS1-fusion-positive lung adenocarcinomas along with immunohistochemistry and FISH assay.
Materials and methods
Study Design and Sample Selection
We performed simultaneous screening of ALK, ROS1, and RET fusions in 295 lung adenocarcinoma specimens by direct, digital transcript profiling using the NanoString’s nCounter technology, as described in a previous study.16 In addition, we collected 500 surgically resected lung adenocarcinoma samples from the Samsung Medical Center (SMC) with previous full informed consent from the patient and with approval from SMC. A total of 795 cases were screened for ALK, EGFR, and KRAS mutation status. Of them, we screened for the presence of RET and ROS1 fusion transcripts in 94 cases that were negative for ALK fusion and also wild type for EGFR and KRAS. We retrospectively reviewed clinicopathologic data. Histologic subtypes of lung adenocarcinoma were classified according to the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society (IASLC/ATS/ERS) multidisciplinary classification of lung adenocarcinoma. We recorded the predominant histologic pattern (lepidic, acinar, papillary, micropapillary, and solid), which can be associated with prognosis. To investigate the association between fusion genes and histologic features, hematoxylin-and-eosin slides were reviewed by two pathologists (YLC and SEL).
RET and ROS1 Immunohistochemistry
Human tissues obtained were fixed in 10% formalin solution, dehydrated through a graded ethanol series, cleared in xylene, and processed for embedding in paraffin wax, according to routine protocols. The sections were incubated in a solution of 0.3% H2O2 for 15 min to inhibit endogenous peroxidase activity. The sections were then incubated for 1 h at RT with primary antibody solutions: RET antibody (ab134100, Abcam, Cambridge, UK, 1:200 dilution) and ROS1 antibody (no. 3287, Cell Signaling Technology, Danvers, MA, USA, 1:40 dilution). The detection systems EnVision+ for Rabbit antibodies (K4003, DAKO, Glostrup, Denmark) were applied according to the manufacturers’ instructions. Slides were stained with liquid diaminobenzidine tetrahydrochloride (DAB+), a high-sensitivity substrate–chromogen system (K3468, DAKO). Counterstaining was performed with Meyer’s hematoxylin. The images on the slides were visualized with an Olympus BX40 light microscope.
Fluorescence In Situ Hybridization
RET and ROS1 FISH tests were performed on formalin-fixed paraffin-embedded (FFPE) tumor tissues using ZytoLight SPEC ROS1 and RET Dual Color Break Apart Probes according to the manufacturer’s instructions (ZytoVision, Bremerhaven, Germany). The SPEC RET Dual Color Break Apart Probe is a mixture of two direct labeled probes hybridizing to the 10q11.21 band. The orange fluorochrome direct labeled probe hybridizes proximal to the RET gene, the green fluorochrome direct labeled probe hybridizes distal to the gene. The SPEC ROS1 Dual Color Break Apart Probe contains green-labeled polynucleotides (ZyGreen: excitation at 503 nm and emission at 528 nm, similar to FITC), which target sequences mapping to 6q22.1 proximal to the ROS1 breakpoint cluster, and orange-labeled polynucleotides (ZyOrange: excitation at 547 nm and emission at 572 nm, similar to rhodamine), which target sequences mapping to 6q22.1 distal to the ROS1 breakpoint cluster. Rearrangement-positive cells were defined as having two rearrangement-positive patterns. One was a break-apart pattern with one fusion signal and two separated green and orange signals (1F1G1O). Only signals that were more than one signal diameter apart from each other were counted as breaks. Another was an isolated 3′ green signal pattern (1F1G). A case was considered positive for rearrangement if >15% of cells showed split signals or single green signals. Signals for each locus-specific FISH probe were assessed under an Olympus BX51TRF microscope (Olympus, Tokyo, Japan) equipped with a triple-pass filter (DAPI/Green/Orange; Vysis, Downers Grove, IL, USA).
RT-PCR and Sequencing
The precise RET/ROS1 fusion variants were determined using RT-PCR, followed by Sanger sequencing. The RNA UltraSense one-step RT-PCR kit (Life Technologies, Carlsbad, CA, USA) was used to generate RT-PCR products. First-strand cDNA was initially synthesized using gene-specific primers.16 cDNA was subdivided into different PCR reactions using the appropriate fusion variant primers, and PCR products were separated on a 2% E-Gel SizeSelect agarose gel (Invitrogen, Carlsbad, CA, USA). In reactions producing a PCR product of the expected size, the amplicons were gel-purified and sequenced using a 3700 ABI Prism sequencer (Applied Biosystems, Foster City, CA, USA).
Statistical Analyses
The χ2 test or Fisher’s exact test was used to examine associations between gene fusion status and clinicopathologic parameters. Overall survival was calculated from the date of diagnosis to the date of death or last follow-up. Recurrence-free survival was defined from the day of first surgery until tumor progression, death, or end of follow-up. Survival analysis was estimated using the Kaplan–Meier method and compared between two or more groups of patients using the log-rank test. Univariate analysis was performed, and the significance of differences in survival between the groups was determined using the log-rank test. Cumulative survival curves and overall survival for groups were computed according to the Kaplan–Meier method. Statistical Package for the Social Sciences (SPSS, Chicago, IL, USA) version 18.0 was used for all statistical analyses. All tests were two-sided, with 0.05 serving as the level of significance.
Results
Clinical Characteristics
RET and ROS1 fusions were found in 15 (16%) and 9 (10%) of 94 EGFR−/KRAS−/ALK− (triple-negative) patients, respectively. The clinical data of the 15 RET fusion-positive and 9 ROS1 fusion-positive patients compared with 70 patients with quintuple-negative (EGFR−/KRAS−/ALK−/ROS1−/RET−) lung adenocarcinoma were summarized in Tables 1, 2, 3. Patients with RET fusion-positive tumors had an younger (P=0.002) median age of 55 years (range, 22–69 years) compared with patients with quintuple-negative lung adenocarcinoma whose median age was 64 years (range, 37–79 years). RET rearrangements were not significantly associated with sex (P=0.492). RET fusion-positive patients had a higher number of never-smokers than quintuple-negative patients (P=0.010). All tumors with RET fusion-positive tumors were classified into the early T stage, such as T1 or T2; however, we found no significant differences between nodal distributions among RET fusion-positive and -negative patients. All but one patient received standard lobectomy and lymph node dissection, the remaining one patient to be treated by wedge resection due to the presence of brain metastasis, with evidence of pathologic stage I in 53%, stage II in 7%, stage III in 33%, and stage IV in 7%. Preoperative chemotherapy and/or radiotherapy were administered to two patients (13%), and seven patients (47%) received postoperative adjuvant therapy. The median follow-up duration was 30 months (range, 2–135 months) after the operation. Of 15 patients, recurrence occurred in four patients (27%) and four patients (27%) died of the disease.
Patients with ROS1 fusion-positive tumors had a median age of 57 years (range, 43–77 years), and 67% of the patients were female; however, there was no statistically significant difference in age and sex. ROS1 fusion-positive patients had a higher number of never-smokers than quintuple-negative patients (P=0.004). ROS1 rearrangements were not significantly associated with T status, N status, and AJCC stage, compared with quintuple-negative patients. All patients received standard lobectomy and lymph node dissection, with evidence of pathologic stage I in 56%, stage II in 22%, stage III in 22%, and stage IV in 0%. Preoperative chemotherapy and/or radiotherapy were administered to one patient (11%), and five patients (56%) received postoperative adjuvant therapy. The median follow-up duration was 38 months (range, 9–53 months) after the operation. Of nine patients, recurrence occurred in two patients (22%) and one patient (11%) died of the disease.
Patients harboring RET/ROS1 fusion have significantly longer RFS than those with quintuple-negative status (P=0.038, P=0.037, respectively). There was no significant difference in overall survival between the fusion-positive and -negative patients (P=0.887). Kaplan—Meier survival curves and corresponding P-values are shown in Figure 1.
Kaplan–Meier survival curves with log-rank test of overall survival (OS) and recurrent-free survival (RFS) according to the RET/ROS1 fusion status. (a) RET/ROS1 fusion had significantly longer RFS than those with quintuple-negative status (P=0.038, P=0.037, respectively). (b) There was no significant difference in OS between the fusion-positive and -negative patients (P=0.887).
Histologic Characteristics
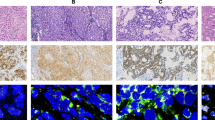
All 15 RET-rearranged tumors showed adenocarcinoma on histology (Table 4). One case (7%) was well differentiated, eight (53%) were moderately differentiated, and six (40%) were poorly differentiated. The predominant growth patterns were acinar in six (40%) cases, papillary in three (20%) cases, solid (Figure 2d) in six (40%) cases. Focal lepidic, solid, and micropapillary component were identified in one, two, and four cases, respectively. Furthermore, psammomatous calcification was seen in three cases. Interestingly, as previously mentioned in the histology of ALK- and ROS1-rearranged lung cancer,16 the solid signet-ring cell pattern (solid growth pattern containing signet-ring cells; Figure 2a) was at least focally present in four (27%) cases and the mucinous cribriform pattern (cribriform structure associated with abundant extracellular mucus; Figures 2b and g) was identified at least focally in four (27%) cases. The solid signet-ring cell pattern was present in three of six (50%) KIF6B-RET-positive tumors and the mucinous cribriform pattern was present in four of five (80%) CCDC6-RET-positive tumors.
Representative RET-rearranged lung adenocarcinomas of case no. 8 (a–c), case no. 9 (d–f), and case no. 10 (g–i). (a) Histologic features of lung adenocarcinoma harboring RET rearrangement. Solid signet-ring cell pattern in hematoxylin-and-eosin (H&E; × 200). (b) Immunohistochemistry of RET, granular cytoplasmic staining pattern ( × 400). (c) Fluorescence in situ hybridization analysis (FISH) pattern of case no. 8. The predominant pattern was 1F2G1O; it consisted of one fusion signal and two separated green and one orange signals. (d) Solid pattern in H&E slides ( × 200). (e) Cytoplasmic and perinuclear aggregate staining pattern in immunohisochemistry ( × 400). (f) FISH pattern of case no. 9. The predominant pattern was 1F1G1O, and the split signal was wide and easily discernible. (g) Mucinous cribriform pattern in H&E slides ( × 200). (h) Granular cytoplasmic staining pattern in immunohisochemistry ( × 400). (i) FISH pattern of case no. 10. The predominant pattern was 1F1G1O, and the split signal was narrow but easily discernible.
All nine ROS1-rearranged tumors also showed adenocarcinoma on histology (Table 5). Seven (78%) cases were moderately differentiated, and one (22%) was poorly differentiated. The predominant growth patterns were acinar in four (44%) cases, papillary in three (33%) cases, and solid in two (22%) cases. Focal solid and micropapillary components were identified in one, and one case, respectively. In addition, psammomatous calcification was seen in one case. As expected, the solid signet-ring cell pattern and mucinous cribriform pattern (Figures 3a and d) were identified at least focally in one (11%) and 3 (33%) cases, respectively. Interestingly, these patterns were present in all four cases of EZR-ROS1-positive tumors.
Representative ROS1-rearranged lung adenocarcinomas of case no. 3 (a–c) and case no. 8 (d–f). (a) Histologic features of lung adenocarcinoma harboring ROS1 rearrangement, mucinous cribriform pattern in hematoxylin-and-eosin (H&E; × 200). (b) Immunohisochemistry of ROS1, cytoplasmic pattern ( × 400). (c) FISH pattern of case no. 3. The predominant pattern was 2G1O and it consisted of two separated green and one orange signals. (d) Mucinous cribriform pattern in H&E ( × 200). (e) cytoplasmic pattern in immunohisochemistry ( × 400). (f) FISH pattern of case no. 8. The predominant pattern was 1F1G and it consisting of an isolated 3′ green signal pattern with one normal fusion signal and one green signal without the corresponding orange signal.
Identification of Fusion Partner Genes
RT-PCR followed by DNA sequencing showed that five tumors carried fusions of KIF5B exon 15 to RET exon 12; and one carried fusion of KIF5B exon 24 to RET exon 12; and five carried fusions of CCDC6 exon 1 to RET exon 12 (Figure 4). One tumor carried fusion of CUX1 exon 10 to RET exon 12, which was recently identified as an additional novel fusion partner gene in a previous study.16 Four tumors carried EZR exon 10-ROS1 exon 34 fusion; three carried SLC34A2 exon 13-ROS1 exon 32 fusions; and one carried CD74 exon 6-ROS1 exon 34 fusion.
Immunohisochemistry Analysis of RET/ROS1 Fusion-Positive Lung Cancer
We performed immunohisochemistry for RET and ROS1 protein expression in 94 triple-negative cases including 24 fusion-positive cases (15 RET+ and 9 ROS1+). Immunohisochemistry data were categorized by the following staining scores: 0=negative, 1=weak, 2=moderate, and 3=strong. In addition, staining pattern was evaluated.
All 15 RET fusion-positive cases showed RET-positive staining but no immunoreactivity for ROS1 protein expression. Of the 15 RET fusion-positive cases, three were scored as moderate and 12 as strong. RET localized diffusely to the cytoplasm in all cases. Both cytoplasmic and membranous patterns were observed in 2 of 15 (13%) cases; the granular cytoplasmic pattern (Figures 2b and h) were observed in 5 of 15 (33%). In one case (7%), cytoplasmic and strong perinuclear aggregates pattern (Figure 2e) similar to the previously observed in ROS1-rearranged adenocarcinoma. On the other hand, 69 of 79 RET fusion-negative cases were completely negative for RET. RET positivity was seen in 10/79 (12%) cases but immunoreactivity extent in all of them was focal (10–30%; Figure 5).
All nine ROS1 fusion-positive cases showed ROS1-positive staining but no immunoreactivity for RET protein expression. Of the nine ROS1 fusion-positive cases, two were scored as moderate and seven as strong. ROS1 localized diffusely to the cytoplasm in all cases (Figures 3b and e); both cytoplasmic and strong punctate staining was observed in one of the nine (11.1%) case. Similar to RET protein staining, normal adjacent tissue did not stain.
FISH Analysis of RET/ROS1 Fusion-Positive Lung Cancer
RET and ROS1 rearrangements were identified in 14 RET-positive tumors (93%) and 7 ROS1-positive tumors (78%), respectively. The FISH probe did not hybridize in the remaining three cases.
The positive RET rearrangement signals (Figures 2c, f and i) ranged from 20 to 97%. The majority of RET rearrangement showed two signal patterns such as one fusion signal and two separated green and orange signals (1F1G1O) and one fusion signal and an isolated 3′ green signal pattern (1F1G). The most common rearrangement signal pattern was the 1F1G1O pattern that was observed in eight cases, and all cases predominantly showed this pattern. All but one case showed a narrow distance between two separated green and orange signals. Notably, in case no. 9 with CUX1 of a novel fusion partner of RET, the split signal was wide and easily discernible. The tumor cells showed the 1F1G rearrangement signal pattern in six cases, and this pattern was predominantly identified in four cases. In one case, an isolated orange signal with a fused signal (1F1O or 2F1O) was identified. The RET copy number was euploidy in the majority of cases; however, copy number gain was seen in seven cases.
The positive ROS1 rearrangement signals (Figures 3c and f) ranged from 50 to 99%. The majority of ROS1 rearrangement also showed two signal patterns such as one fusion signal and two separated green and orange signals (1F1G1O) and an isolated 3′ green signal pattern with one normal fusion signal and one green signal without the corresponding green signal (1F1G). The most common rearrangement signal pattern was the 1F1G1O pattern observed in four cases, and two out of 4the four cases predominantly showed this pattern. Regardless of the fusion partner genes, the distance of two separated green and orange signals was wide and easily discernible in all cases. The tumor cells showed a 1F1G rearrangement signal pattern in four cases, and two out of the four cases, this pattern was predominantly identified. ROS1 copy number gain was seen in four cases.
Discussion
RET was mapped to chromosome 10q11.2, where it encodes a RTK.17 Chromosomal rearrangements involving the RET proto-oncogene in papillary thyroid cancer were reported in 1990.18 CCDC6-RET18 and NCOA4-RET19 rearrangements account for the majority of radiation induced and sporadic papillary thyroid cancers.20 Since the initial report of RET rearrangements in lung adenocarcinoma by Ju et al10 in 2012, about 100 cases have been described in the literature.6, 21, 22 Our study revealed that 16% (15 out of 94) of EGFR−/KRAS−/ALK− (triple-negative) lung adenocarcinomas harbored RET rearrangement. This prevalence was higher than that of the entire adenocarcinoma cohort because we screened the triple-negative cohort. RET rearrangements are mutually exclusive with other oncogenic alterations such as EGFR, KRAS, ALK, ERBB2, and BRAF, suggesting that RET fusions are independent oncogenic drivers in lung cancer. Although fusion genes are oncogenic drivers, they present in lung cancer at low frequency. Therefore, identifying the enriched population of fusion genes in lung cancer could contribute to future clinical screening.
Current methods for the detection of ALK, ROS1, and RET fusions are FISH, immunohisochemistry, and/or RT-PCR, each assay having its own advantages and disadvantages. However, it is difficult to screen all genomic alterations using these tools in routine clinical practice. Thus, recognizing distinctive clinicopathologic features, especially histologic features, may help to find candidates to screen for genomic alterations. Some studies recently reported the clinicopathologic features of RET rearrangements.15, 21, 22 Our study also showed similar results in that patients with RET fusion-positive tumors were younger in age (median age, 55 vs 64 years; P=0.002), never-smokers (P=0.010) and early T stage (P<0.001) compared with patients with RET fusion-negative lung adenocarcinoma. However, there was no statistically significant difference in the N stage.
Interestingly, as previously reported as a histopathological marker for the presence of EML4-ALK13, 23, 24, 25, 26 and as mentioned as histologic features of ROS1-rearranged lung cancer,27 the solid signet-ring cell pattern and mucinous cribriform pattern were identified at least focally in RET fusion cases. As expected, these patterns were observed in ROS1-rearranged lung cancer. Interestingly, the mucinous cribriform pattern was present in four of five (80%) CCDC6-RET-positive tumors and the solid signet-ring cell pattern was present in three of six (50%) KIF6B-RET-positive tumors. Similar to our results, Takeuchi et al13 recently described lung cancer harboring a CCDC6-RET rearrangement with a mucinous cribriform pattern. Although further study focusing on the relation between these histologic features and the fusion partner genes of RET rearrangement is needed, an important current finding was that fusion gene-associated lung cancer shares similar histologic features. When pathologists encounter an adenocarcinoma showing these histologic patterns in routine histopathological diagnosis, it could be helpful to test for the gene fusion. Therefore, recognizing the characteristic histologic features of fusion gene-associated lung cancer is critical.
We detected 15 RET fusion transcripts in the previous study and confirmed the fusion status using RT-PCR followed by sequencing. Currently, five fusion partners to RET (KIF5B, CCDC6, TRIM33, NCOA4, and CUX1) have been identified in lung cancer.15, 28, 29 All of these genes are located on chromosome 10 except TRIM33 and CUX1. Translocations can occur within a chromosome (intrachromosomal) or between chromosomes (interchromosomal). KIF5B is the most common fusion partner in lung cancer, and our study showed similar results. Therefore, RET fusion in lung adenocarcinoma appears to arise predominantly through intrachromosomal rearrangement. In our study, the majority of RET rearrangements showed narrow split with a distance approaching the diameter of one to two hybridization signals. However, a case of exon 10 of the CUX1 gene (7q22.1) fusing to exon 12 of the RET gene (10q11.21) showed a wide split signal. This finding suggests that RET FISH analysis showed differences in interchromosomal and intrachromosomal translocations. The KIF5B (10p11.22) and CCDC6 (10q21.2) genes are located in the same chromosome from the RET gene. In this situation, the simplest mechanism to generate fusion would be interstitial deletion that would result in an FG FISH pattern.27 However, the FGO pattern was predominantly shown in these intrachromosomal translocation cases. Therefore, it does not seem to be enough to explain the differences in the FGO and FG FISH patterns. Yoshida et al27 recently reported no association between ROS1 fusion partners and FISH signal patterns. We also showed that there were no differences in the FGO and FG FISH patterns and no differences in intra- and interchromosomal translocations in ROS1 FISH analysis. It is important to consider a negative control, such as normal non-neoplastic lung tissues, for establishing a cutoff value for accurate FISH. We observed that all of the RET/ROS1 FISH-positive cases showed abnormal split or isolated green signals in more than 20% of tumor cells. We performed RET FISH on the nine ROS1-positive confirmed cases and ROS1 FISH on the 15 RET-positive confirmed cases. In both experiments, the split was found in less than 3% of tumor cells. The performed 10 quintuple-negative cases showed less than 3% of split signals in both RET and ROS1 FISH test. From our experiment, the cutoff value should be between 3 and 20%. From the referred ALK study, we have chosen the cutoff of 15% for both RET and ROS1 FISH tests. Although break-apart FISH is currently the most effective diagnostic tool to detect chromosomal rearrangements, it has not been used routinely in clinical practice because of the high cost and need for technical expertise. Moreover, specific and unknown variants of fusion genes cannot be distinguished by the break-apart FISH assay.
Historically, ROS1 immunohistochemistry was an ineffective tool in the diagnosis of fusion status because ROS1 mRNA was known to be overexpressed in 20–30% of lung adenocarcinomas30 regardless of gene rearrangement status.31 However, a novel ROS1 immunohistochemistry assay has been developed recently, with no ROS1 staining in both adjacent normal lung tissue and wild-type lung cancer using the D4D6 antibody.32 Furthermore, additional studies have validated ROS1 immunohistochemistry using the D4D6 antibody and the results are suggestive that ROS1 immunohistochemistry may be an effective screening tool.6, 33, 34 In our study, all nine ROS1 fusion-positive cases showed positivity of more than moderate intensity, for the ROS1 protein. Similarly, RET immunohistochemistry has been ineffective as a screening tool because some studies showed no significant differences between RET immunohistochemical staining patterns among RET-positive and RET-negative specimens.12, 15, 35 However, in our study, all 15 RET fusion-positive cases showed positivity of more than moderate intensity for the RET protein, and the majority showed cytoplasmic staining. On the other hand, in 79 RET fusion-negative cases, 69 cases did not stain at all. Benchmarking to NanoString’s nCounter screening results, RET immunohistochemistry is 100% sensitive and 87.3% specific for the presence of RET fusion. Comparing with the ALK immunohistochemistry, which is a reliable screening tool for the identification of ALK rearrangement, ALK immunohistochemistry assay showed 66–100% sensitivity and 63–100% specificity using various antibody systems.36 Therefore, we recommend RET and ROS1 immunohistochemistry as a possible adjunctive diagnostic tool for the detection of RET and ROS1 rearrangements in FFPE tissues.
To the best of our knowledge, this is the largest study to describe detailed histological findings and FISH patterns in RET rearrangement lung cancer. Testing the fusion status of the ROS1 and RET genes should be considered in select patients, such as those with adenocarcinoma histology together with aforementioned fusion-associated histologic features (solid signet-ring cell pattern and mucinous cribriform pattern).
In conclusion, our study has provided characteristic fusion gene-associated histologic features. We further proposed future screening strategies and enabled clinicians to direct patients to clinical trials targeting such populations.
References
Soda M, Choi YL, Enomoto M et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561–566.
Kwak EL, Bang YJ, Camidge DR et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693–1703.
Shaw AT, Yeap BY, Mino-Kenudson M et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 2009;27:4247–4253.
Saijo N . Present status and problems on molecular targeted therapy of cancer. Cancer Res Treat 2012;44:1–10.
Rikova K, Guo A, Zeng Q et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190–1203.
Gainor JF, Shaw AT . Novel targets in non-small cell lung cancer: ROS1 and RET fusions. Oncologist 2013;18:865–875.
Bergethon K, Shaw AT, Ou SH et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863–870.
Chin LP, Soo RA, Soong R et al. Targeting ROS1 with anaplastic lymphoma kinase inhibitors: a promising therapeutic strategy for a newly defined molecular subset of non-small-cell lung cancer. J Thorac Oncol 2012;7:1625–1630.
McDermott U, Iafrate AJ, Gray NS et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res 2008;68:3389–3395.
Ju YS, Lee WC, Shin JY et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res 2012;22:436–445.
Kohno T, Ichikawa H, Totoki Y et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012;18:375–377.
Lipson D, Capelletti M, Yelensky R et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 2012;18:382–384.
Takeuchi K, Soda M, Togashi Y et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378–381.
Cai W, Su C, Li X et al. KIF5B-RET fusions in Chinese patients with non-small cell lung cancer. Cancer 2013;119:1486–1494.
Wang R, Hu H, Pan Y et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol 2012;30:4352–4359.
Lira ME, Choi YL, Lim SM et al. A single-tube multiplexed assay for detecting ALK, ROS1, and RET fusions in lung cancer. J Mol Diagn 2014;16:229–243.
Ishizaka Y, Itoh F, Tahira T et al. Human ret proto-oncogene mapped to chromosome 10q11.2. Oncogene 1989;4:1519–1521.
Grieco M, Santoro M, Berlingieri MT et al. PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell 1990;60:557–563.
Santoro M, Dathan NA, Berlingieri MT et al. Molecular characterization of RET/PTC3; a novel rearranged version of the RETproto-oncogene in a human thyroid papillary carcinoma. Oncogene 1994;9:509–516.
Nikiforov YE, Nikiforova MN . Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol 2011;7:569–580.
Tsuta K, Kohno T, Yoshida A et al. RET-rearranged non-small-cell lung carcinoma: a clinicopathological and molecular analysis. Br J Cancer 2014;110:1571–1578.
Pan Y, Zhang Y, Li Y et al. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: A comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer 2014;84:121–126.
Jokoji R, Yamasaki T, Minami S et al. Combination of morphological feature analysis and immunohistochemistry is useful for screening of EML4-ALK-positive lung adenocarcinoma. J Clin Pathol 2010;63:1066–1070.
Rodig SJ, Mino-Kenudson M, Dacic S et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res 2009;15:5216–5223.
Yoshida A, Tsuta K, Nakamura H et al. Comprehensive histologic analysis of ALK-rearranged lung carcinomas. Am J Surg Pathol 2011;35:1226–1234.
Ha SY, Ahn J, Roh MS et al. Cytologic features of ALK-positive pulmonary adenocarcinoma. Korean J Pathol 2013;47:252–257.
Yoshida A, Kohno T, Tsuta K et al. ROS1-rearranged lung cancer: a clinicopathologic and molecular study of 15 surgical cases. Am J Surg Pathol 2013;37:554–562.
Chao BH, Briesewitz R, Villalona-Calero MA . RET fusion genes in non-small-cell lung cancer. J Clin Oncol 2012;30:4439–4441.
Drilon A, Wang L, Hasanovic A et al. Response to Cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov 2013;3:630–635.
Acquaviva J, Wong R, Charest A . The multifaceted roles of the receptor tyrosine kinase ROS in development and cancer. Biochim Biophys Acta 2009;1795:37–52.
Li C, Fang R, Sun Y et al. Spectrum of oncogenic driver mutations in lung adenocarcinomas from East Asian never smokers. PLoS ONE 2011;6:e28204.
Rimkunas VM, Crosby KE, Li D et al. Analysis of receptor tyrosine kinase ROS1-positive tumors in non-small cell lung cancer: identification of a FIG-ROS1 fusion. Clin Cancer Res 2012;18:4449–4457.
Sholl LM, Sun H, Butaney M et al. ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am J Surg Pathol 2013;37:1441–1449.
Mescam-Mancini L, Lantuejoul S, Moro-Sibilot D et al. On the relevance of a testing algorithm for the detection of ROS1-rearranged lung adenocarcinomas. Lung Cancer 2014;83:168–173.
Sasaki H, Shimizu S, Tani Y et al. RET expression and detection of KIF5B/RET gene rearrangements in Japanese lung cancer. Cancer Med 2012;1:68–75.
Conklin CM, Craddock KJ, Have C et al. Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J Thorac Oncol 2013;8:45–51.
Acknowledgements
This work was supported by a grant of the Korea Healthcare Technology R&D Project, Ministry for Health and Welfare Affairs, Republic of Korea (A092255), and the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science, ICT and Future Planning (MSIP; NRF-2013R1A2A2A01068922).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
ME Lira is an employee of Pfizer, Inc. The remaining authors declare no conflicts of interest.
Rights and permissions
About this article
Cite this article
Lee, S., Lee, B., Hong, M. et al. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod Pathol 28, 468–479 (2015). https://doi.org/10.1038/modpathol.2014.107
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2014.107
This article is cited by
-
Rare molecular subtypes of lung cancer
Nature Reviews Clinical Oncology (2023)
-
Clinicopathological features and prognostic significance of pulmonary adenocarcinoma with signet ring cell components: meta-analysis and SEER analysis
Clinical and Experimental Medicine (2023)
-
Bronchial salivary gland–type intraductal carcinoma with KIAA1217::RET gene fusion composed of intercalated and oncocytic components
Virchows Archiv (2023)
-
Anwendungen der FISH in der Diagnostik von Lungenkarzinomen
Wiener klinisches Magazin (2021)
-
Anwendungen der FISH in der Diagnostik von Lungenkarzinomen
Der Pathologe (2020)