Abstract
A recurrent intrachromosomal rearrangement on chromosome 12q in solitary fibrous tumor leads to the formation of a NAB2–STAT6 fusion oncogene. As a result, nuclear expression of the cytoplasmic transcription factor STAT6 is found in solitary fibrous tumor and serves as a useful diagnostic marker. STAT6 is located in 12q13, a region containing well-characterized oncogenes that are commonly amplified in dedifferentiated liposarcoma; we have previously reported that STAT6 is expressed in a subset of dedifferentiated liposarcoma. The aim of this study was to determine the frequency of STAT6 expression in dedifferentiated liposarcoma and the underlying genetic mechanism. STAT6 protein expression was analyzed by immunohistochemistry in a well-characterized series of 35 previously unpublished cases of dedifferentiated liposarcoma, all with nuclear MDM2 and/or CDK4 expression by immunohistochemistry and/or cytogenetic features of dedifferentiated liposarcoma. FISH for STAT6 was performed in all cases with STAT6 expression, and a subset of control cases without STAT6 expression. In total 4/35 cases (11%) showed STAT6 expression (three with multifocal staining of moderate to strong intensity and one with weak focal staining). FISH demonstrated amplification of STAT6 in all cases positive for STAT6 by immunohistochemistry; in contrast, FISH performed on four STAT6-negative dedifferentiated liposarcomas demonstrated no STAT6 amplification (P=0.0286). Of the four STAT6 amplified cases, three patients were male and one was female, ranging in age from 51 to 76 years. Tumors were located in the mediastinum (n=2), paratesticular soft tissue (n=1), and perirenal soft tissue (n=1). Three patients received pre-operative chemotherapy +/− radiation therapy. In conclusion, STAT6 is amplified in a subset of dedifferentiated liposarcoma, resulting in STAT6 protein expression that can be detected by immunohistochemistry and may be a potential pitfall in the differential diagnosis of dedifferentiated liposarcoma and solitary fibrous tumor. These findings suggest a role for STAT6-mediated transcriptional activity in some cases of dedifferentiated liposarcoma and highlight the genomic complexity and heterogeneity of dedifferentiated liposarcoma.
Similar content being viewed by others
Main
Liposarcoma is the most common sarcoma in adults. Well-differentiated and dedifferentiated liposarcoma account for 40–45% of all liposarcomas and fall within a biological continuum in which dedifferentiated liposarcoma represents an advanced form of the tumor that has acquired metastatic potential.1 Clinically, dedifferentiated liposarcoma is an aggressive malignancy that is frequently lethal due to multiple recurrences and late metastases, with no effective therapeutic options other than surgery.1, 2, 3 Morphologically, dedifferentiated liposarcoma is usually a non-lipogenic high-grade sarcoma with a wide range of histological appearances, most commonly with spindle cell and pleomorphic cytomorphology, and infrequently inflammatory, round cell, or meningothelial-like patterns, none of which seem to correlate with the clinical outcome.1, 2
Cytogenetic and molecular genetic studies over the last two decades have provided insights into the biology of dedifferentiated liposarcoma. The tumor cells of dedifferentiated liposarcoma contain neochromosomes —ring or giant marker chromosomes— composed of repeated interspersed amplicons from distinct non-contiguous chromosomal regions.4, 5 The structure of dedifferentiated liposarcoma neochromosomes is complex and varies between tumors, between time points in the course of disease, and even between individual cells within a single tumor. Despite such genetic heterogeneity, some general features are observed in dedifferentiated liposarcoma neochromosomes: genetic material from the 12q13-15 chromosomal region is invariably present, usually as small fragments of variable lengths that are amplified at very high levels. Other chromosomes are involved less consistently, such as 1q, 4p, 12q21–22, 13q, and 15q.5, 6, 7, 8 The 12q15 amplicon consistently contains the MDM2 oncogene, which is considered an essential oncogenic driver in dedifferentiated liposarcoma and the main downregulator of the p53 axis in this tumor type.1, 9 Concomitant amplification of HMGA2, CPM, FRS2, and YEATS4, all located at the same chromosomal region, is invariably present along with MDM2, even though the pathogenetic relevance of these genes in dedifferentiated liposarcoma is not well established.5, 10, 11, 12 In addition, the cyclin-dependent kinase CDK4 is amplified in about 90% of dedifferentiated liposarcoma. Although CDK4 is located very close to the main 12q15 amplicon, it belongs to a discontinuous and inconsistent amplicon that includes its locus at 12q14.1 as well as other genes such as TSPAN31.6, 10 CDK4 gene amplification and overexpression very likely have an important role in the tumorigenic process in dedifferentiated liposarcoma, but the contribution of other genes in this amplicon has not been explored.13, 14 In addition to providing greater understanding of the pathogenesis of dedifferentiated liposarcoma, the concurrent amplification and overexpression of MDM2 and CDK4 serve as important diagnostic markers that can be detected by immunohistochemistry or FISH.15 MDM2 and CDK4 also represent attractive potential therapeutic targets—both MDM2 inhibition and CDK4 inhibition have shown promising activity in preclinical models and are being evaluated in clinical trials.3, 16
STAT6 is a member of the STAT family of cytoplasmic transcription factors, which regulate gene expression by transmitting signals to the nucleus and binding to specific DNA promoter sequences in response to extracellular cytokines. Activation of STAT family members has been implicated in physiological and neoplastic cell growth, and can be therapeutically inhibited.17 In mesenchymal neoplasia, recent studies have detected a recurrent intrachromosomal rearrangement in solitary fibrous tumor that results in the formation of a NAB2–STAT6 fusion oncogene.18, 19 The fusion oncoprotein is highly expressed and localizes to the cell nucleus, resulting in a strong nuclear expression of STAT6 that can be detected by immunohistochemistry and serves as a reliable diagnostic marker for solitary fibrous tumor.20, 21 In addition, we have previously reported that a small number of dedifferentiated liposarcomas show STAT6 expression.20 STAT6 is located at chromosomal region 12q13, which contains essential oncogenes amplified in dedifferentiated liposarcoma neochromosomes. The aim of this study was to determine the frequency of STAT6 expression in dedifferentiated liposarcoma and the underlying genetic mechanism, in relationship to the known amplification of 12q13-15 in this sarcoma type.
Materials and methods
The study was approved by the Institutional Review Board at Brigham and Women’s Hospital. Thirty-five previously unpublished cases of dedifferentiated liposarcoma were retrieved from the surgical pathology files of Brigham and Women’s Hospital, Boston, MA, USA. Clinical, histopathological, and immunophenotypical features were reviewed by two surgical pathologists (LAD and AME). All cases showed nuclear expression of MDM2 and CDK4 by immunohistochemistry, and/or cytogenetic features of dedifferentiated liposarcoma—either ring or giant marker chromosomes in the karyotype, MDM2 amplification by FISH, or both. Two solitary fibrous tumors with STAT6 overexpression were selected as controls.
Immunohistochemistry for STAT6 was performed on 4 μm-thick formalin-fixed paraffin-embedded whole-tissue sections following pressure cooker antigen retrieval (0.01 M citrate buffer; pH, 6.0), using a rabbit polyclonal antibody directed against the C-terminus of STAT6 (1:1000; sc-621; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Appropriate positive and negative controls were used throughout. The extent of immunoreactivity was graded according to the percentage of positive tumor cells (0, no staining; 1+, 1–50%; 2+, 51–100%). The intensity of staining was graded as weak, moderate, or strong.
Dual color FISH was performed on 4 μm thick formalin-fixed paraffin-embedded tissue sections for all cases with STAT6 expression, a subset of four dedifferentiated liposarcoma cases without STAT6 expression, and two solitary fibrous tumor controls. Tissue sections were baked overnight at 56 °C, deparaffinized by three consecutive 15 min immersions in xylene, and dehydrated twice in 100% ethanol for 2 min. The slides were then immersed in TRIS–EDTA (100 mM Tris base, 50 mM EDTA, pH 7.0) for 1 h at 95–99 °C and rinsed in PBS for 5 min. Proteolytic digestion of the sections was performed using Digest–All 3 Pepsin solution (Invitrogen, Camarillo, CA, USA) at 37 °C for 10–20 min, twice. The sections were then sequentially dehydrated in alcohol (70, 85, 95, and 100% for 2 min each) and air-dried. The STAT6 probe set consisted of a commercial kit targeting the 12q13 region, which includes a SpectrumOrange probe spanning the STAT6 locus and a telomeric SpectrumGreen probe that spans the CDK4 locus (Vysis, Abbott Molecular Inc, Des Plaines, IL, USA) (Figure 1). One-hundred interphase nuclei were scored per case. The presence of five or more orange signals was considered indicative of STAT6 amplification. Non-neoplastic diploid cells were identified in each case and used as internal controls. The significance of the correlation between immunohistochemistry and FISH was analyzed using a 2 × 2 contingency table, and the exact two-tailed P-value was calculated according to Fisher’s test.
Results
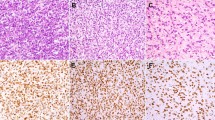
Four of the 35 dedifferentiated liposarcomas (11%) showed STAT6 expression by immunohistochemistry; the clinicopathologic features of these cases are summarized in Table 1. Morphologically, two tumors were high-grade spindle cell and pleomorphic sarcomas (one with focal heterologous smooth muscle differentiation), one was composed of a relatively monotonous population of atypical spindle cells with prominent nuclear palisading, and one was a high-grade spindle-cell sarcoma with hypocellular hyalinized areas characteristic of well-differentiated sclerosing liposarcoma. Three cases showed multifocal (2+) staining of moderate to strong intensity and one (case 4) showed weak focal (1+) staining (Figure 2). STAT6 expression was both cytoplasmic and nuclear in three cases, generally with a stronger nuclear intensity; in case one, although some areas of the tumor predominantly showed nuclear staining, cytoplasmic staining was also present multifocally. STAT6 expression was detected in the dedifferentiated components in all four tumors, and in an area of well-differentiated sclerosing liposarcoma in case four. Interestingly, the tumor with heterologous smooth muscle differentiation (case 2) showed expression of STAT6 only in conventional areas of dedifferentiated liposarcoma; the smooth muscle-like component was negative for STAT6 by immunohistochemistry. Two solitary fibrous tumor control cases showed diffuse strong nuclear STAT6 expression.
High-grade dedifferentiated liposarcoma with pleomorphic and spindle-cell morphology (case 1) (a) showing diffuse expression of STAT6 (b). Dedifferentiated liposarcoma with predominantly spindle cell morphology and prominent nuclear palisading (case 3) (c). The tumor cells showed diffuse nuclear expression and weak cytoplasmic expression of STAT6 (d). Hypocellular hyalinized areas of well-differentiated sclerosing liposarcoma (case 4) (e) showed weak focal expression of STAT6 (f).
FISH demonstrated STAT6 amplification in all four cases expressing STAT6. The level of amplification was variable between cases, ranging from high-level amplification with innumerable copies of STAT6 per cell (Figure 3a), to 6–10 STAT6 copies present in one case. Within each case, there were also different levels of amplification in different cells, with large atypical cells showing more gene copies than less atypical cells. The smooth muscle-like component present in case two, identified by elongated nuclei arranged in fascicles on the DAPI stain, showed high-level amplification. FISH performed on four dedifferentiated liposarcomas negative for STAT6 expression demonstrated no STAT6 amplification (P=0.0286). All dedifferentiated liposarcomas showed high-level amplification of CDK4 (Figure 3b). CDK4 amplification was consistently higher than STAT6 amplification. Both STAT6-positive solitary fibrous tumors showed just two copies of STAT6 and CDK4 per interphase nucleus.
STAT6 amplification detected in dedifferentiated liposarcoma interphase nuclei by FISH. A non-neoplastic cell is shown in each field as an internal control. (a) High-level amplification of STAT6 demonstrated by the presence of multiple red signals, isolated, or overlapping with green signals. CDK4 is highly amplified, as represented by innumerable green signals (case 1). (b) No amplification of STAT6 despite high-level amplification of CDK4 in a dedifferentiated liposarcoma control case, negative for STAT6 by immunohistochemistry.
Of the four STAT6 amplified dedifferentiated liposarcomas, three patients were males and one was female, ranging in age from 51 to 76 years (Table 1). Tumors were located in the mediastinum (n=2), paratesticular soft tissue (n=1), and perirenal soft tissue (n=1). Three patients received pre-operative chemotherapy +/− radiation therapy before resection.
Discussion
The significance of STAT6 amplification and overexpression in a subset of dedifferentiated liposarcoma must be considered from both a diagnostic and a biological perspective, with potential therapeutic implications. Diagnostically, the specificity of STAT6 immunodetection for the diagnosis of solitary fibrous tumor in the appropriate morphological context is very high.20, 21 Dedifferentiated liposarcoma, however, can resemble solitary fibrous tumor morphologically, and STAT6 expression in a subset of dedifferentiated liposarcoma could complicate this differential diagnosis. The degree of STAT6 expression in solitary fibrous tumor is generally more diffuse and of a stronger intensity than in dedifferentiated liposarcoma, although the range of expression is variable, limiting the value of this observation for the evaluation of individual cases. The predominantly nuclear aberrant localization of STAT6 in solitary fibrous tumor, in contrast to the cytoplasmic and nuclear expression seen in dedifferentiated liposarcoma, may be a helpful discriminatory feature. Nevertheless, expression of STAT6 in dedifferentiated liposarcoma is a potential diagnostic pitfall that must be taken into consideration when evaluating a STAT6-positive spindle cell neoplasm, particularly in small needle biopsies of retroperitoneal or deep lesions. The presence of areas with varied morphologies, including areas of well-differentiated liposarcoma, and the demonstration of MDM2 and CDK4 expression by immunohistochemistry, or amplification by FISH, should allow for a diagnosis of dedifferentiated liposarcoma in the vast majority of cases.11, 15 However, the mere presence of fat formation within the tumor should not prompt a diagnosis of dedifferentiated liposarcoma, given the existence of a well-recognized subgroup of fat-forming solitary fibrous tumor, both benign and malignant.22, 23
Mechanistically, amplification of STAT6 in dedifferentiated liposarcoma is in keeping with the chromosomal location of this gene at 12q13 and the genetic makeup of dedifferentiated liposarcoma. Consistent MDM2 amplification and frequent CDK4 coamplification highlight the relevance of 12q13-15 amplicons in dedifferentiated liposarcoma,4, 10, 24 but the remarkable heterogeneity of these and other amplicons in dedifferentiated liposarcoma is still poorly understood. Ring and giant marker chromosomes in dedifferentiated liposarcoma are topologically complex structures and are thought to result from multiple rounds of sequential breakage, fusion and bridging of donor chromosomes.25 The resulting neochromosomes show a notable degree of plasticity, as demonstrated by the genetic variability between cases, and within each case.4 Some structural considerations have been proposed to explain the composition of certain dedifferentiated liposarcoma amplicons; for example, a fragile site located immediately centromeric to the 5′ end of DDIT3 at 12q14.1 has been proposed to define the centromeric boundary of the CDK4 amplicon, so that chromosomal breakage at this site would have a role in the formation of the non-contiguous amplicons from 12q13-15.6 Nevertheless, several genes centromeric to this breakpoint are coamplified in around 15% of dedifferentiated liposarcoma (including GLI1, DCTN2,6, 14 and as demonstrated in this report, STAT6), indicating that some cases of dedifferentiated liposarcoma contain either larger amplicons extending over this site, or additional discontinuous amplicons from 12q13.
The relevance of STAT6 amplification in the pathogenesis of dedifferentiated liposarcoma remains to be defined. The relatively low frequency of STAT6 amplification in dedifferentiated liposarcoma indicates that it is most likely a passenger genomic event. Nevertheless, STAT6 may contribute substantially to the biology of dedifferentiated liposarcoma. Evidence in support of this interpretation originates from the only study that systematically analyzed the functional relevance of amplified genes in dedifferentiated liposarcoma: shRNA-mediated STAT6 knockdown in three dedifferentiated liposarcoma cell lines significantly reduced cell proliferation, with lethal effects comparable to or greater than those resulting from CDK4 knockdown.14 Despite recent advances in the characterization of dedifferentiated liposarcoma neochromosomes, the functional contribution of each of the genes amplified within dedifferentiated liposarcoma amplicons is still unclear. Although MDM2 is the best studied target of the 12q15 amplicon,11, 16 the consistent presence within the same amplicon of YEATS4 and FRS2 amplification, combined with HMGA2 rearrangement, have also been demonstrated to be biologically essential.6, 12 Abundant experimental evidence also supports the functional relevance of CDK4 in dedifferentiated liposarcoma.13, 14 Less common genomic events have been variably associated with particular biological features, for example, amplification of JUN and dedifferentiation.8 The presence of STAT6 amplification demonstrated in this report, combined with the functional evidence of STAT6 dependency in dedifferentiated liposarcoma,14 highlights the biological and potentially therapeutic relevance of STAT6 in at least a subset of dedifferentiated liposarcoma and provides a basis to explore STAT6-targeting agents17 as a rational therapeutic strategy in dedifferentiated liposarcoma.
In summary, STAT6 amplification occurs in a subset of dedifferentiated liposarcoma, which results in expression of STAT6 that can be detected by immunohistochemistry and may be a potential pitfall in the differential diagnosis of dedifferentiated liposarcoma and solitary fibrous tumor. STAT6 amplification in dedifferentiated liposarcoma further highlights the genomic complexity and heterogeneity of ring and giant marker chromosomes of this tumor type, particularly concerning amplicons originating from the chromosomal region 12q13-15. STAT6 transcriptional activity may be biologically relevant in some cases of dedifferentiated liposarcoma and may provide a therapeutic opportunity.
References
Fletcher CDM, Bridge JA, Hogendoorn PCW, Mertens F, (eds).. WHO Classification of Tumours of Soft Tissue and Bone 4th edn. IARC: Lyon, France, 2013, pp 468.
McCormick D, Mentzel T, Beham A et al. Dedifferentiated liposarcoma. Clinicopathologic analysis of 32 cases suggesting a better prognostic subgroup among pleomorphic sarcomas. Am J Surg Pathol 1994;18:1213–1223.
Crago AM, Singer S . Clinical and molecular approaches to well differentiated and dedifferentiated liposarcoma. Curr Opin Oncol 2011;23:373–378.
Garsed DW, Holloway AJ, Thomas DM . Cancer-associated neochromosomes: a novel mechanism of oncogenesis. Bioessays 2009;31:1191–1200.
Pedeutour F, Forus A, Coindre JM et al. Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors. Genes Chromosomes Cancer 1999;24:30–41.
Italiano A, Bianchini L, Keslair F et al. HMGA2 is the partner of MDM2 in well-differentiated and dedifferentiated liposarcomas whereas CDK4 belongs to a distinct inconsistent amplicon. Int J Cancer 2008;122:2233–2241.
Persson F, Olofsson A, Sjogren H et al. Characterization of the 12q amplicons by high-resolution, oligonucleotide array CGH and expression analyses of a novel liposarcoma cell line. Cancer Lett 2008;260:37–47.
Snyder EL, Sandstrom DJ, Law K et al. C-jun amplification and overexpression are oncogenic in liposarcoma but not always sufficient to inhibit the adipocytic differentiation programme. J Pathol 2009;218:292–300.
Ray-Coquard I, Blay JY, Italiano A et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol 2012;13:1133–1140.
Pedeutour F, Suijkerbuijk RF, Forus A et al. Complex composition and co-amplification of SAS and MDM2 in ring and giant rod marker chromosomes in well-differentiated liposarcoma. Genes Chromosomes Cancer 1994;10:85–94.
Dei Tos AP, Doglioni C, Piccinin S et al. Coordinated expression and amplification of the MDM2, CDK4, and HMGI-C genes in atypical lipomatous tumours. J Pathol 2000;190:531–536.
Wang X, Asmann YW, Erickson-Johnson MR et al. High-resolution genomic mapping reveals consistent amplification of the fibroblast growth factor receptor substrate 2 gene in well-differentiated and dedifferentiated liposarcoma. Genes Chromosomes Cancer 2011;50:849–858.
Italiano A, Bianchini L, Gjernes E et al. Clinical and biological significance of CDK4 amplification in well-differentiated and dedifferentiated liposarcomas. Clin Cancer Res 2009;15:5696–5703.
Barretina J, Taylor BS, Banerji S et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet 2010;42:715–721.
Binh MB, Sastre-Garau X, Guillou L et al. MDM2 and CDK4 immunostainings are useful adjuncts in diagnosing well-differentiated and dedifferentiated liposarcoma subtypes: a comparative analysis of 559 soft tissue neoplasms with genetic data. Am J Surg Pathol 2005;29:1340–1347.
Conyers R, Young S, Thomas DM . Liposarcoma: molecular genetics and therapeutics. Sarcoma 2011; 2011:483154.
Buettner R, Mora LB, Jove R . Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res 2002;8:945–954.
Robinson DR, Wu YM, Kalyana-Sundaram S et al. Identification of recurrent NAB2-STAT6 gene fusions in solitary fibrous tumor by integrative sequencing. Nat Genet 2013;45:180–185.
Mohajeri A, Tayebwa J, Collin A et al. Comprehensive genetic analysis identifies a pathognomonic NAB2/STAT6 fusion gene, nonrandom secondary genomic imbalances, and a characteristic gene expression profile in solitary fibrous tumor. Genes Chromosomes Cancer 2013;52:873–886.
Doyle LA, Vivero M, Fletcher CD et al. Nuclear expression of STAT6 distinguishes solitary fibrous tumor from histologic mimics. Mod Pathol advance online publication, 13 September 2013 doi:10.1038/modpathol.2013.164(e-pub ahead of print).
Schweizer L, Koelsche C, Sahm F et al. Meningeal hemangiopericytoma and solitary fibrous tumors carry the NAB2-STAT6 fusion and can be diagnosed by nuclear expression of STAT6 protein. Acta Neuropathol 2013;125:651–658.
Lee JC, Fletcher CD . Malignant fat-forming solitary fibrous tumor (so-called ‘lipomatous hemangiopericytoma’): clinicopathologic analysis of 14 cases. Am J Surg Pathol 2011;35:1177–1185.
Guillou L, Gebhard S, Coindre JM . Lipomatous hemangiopericytoma: a fat-containing variant of solitary fibrous tumor? Clinicopathologic, immunohistochemical, and ultrastructural analysis of a series in favor of a unifying concept. Hum Pathol 2000;31:1108–1115.
Nilbert M, Rydholm A, Mitelman F et al. Characterization of the 12q13-15 amplicon in soft tissue tumors. Cancer Genet Cytogenet 1995;83:32–36.
Gisselsson D, Pettersson L, Hoglund M et al. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci U S A 2000;97:5357–5362.
Acknowledgements
We are very grateful to Dr Jonathan A Fletcher for insightful discussions and valuable feedback about this study. AME is supported by a Career Development Award from the Sarcoma Alliance for Research through Collaboration (SARC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Doyle, L., Tao, D. & Mariño-Enríquez, A. STAT6 is amplified in a subset of dedifferentiated liposarcoma. Mod Pathol 27, 1231–1237 (2014). https://doi.org/10.1038/modpathol.2013.247
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.247
Keywords
This article is cited by
-
A case of fat-forming solitary fibrous tumor that is prone to be confused with liposarcoma
Diagnostic Pathology (2024)
-
A successful trimodality therapy for difficult-to-diagnose primary mediastinal dedifferentiated liposarcoma, which originated from the perihilar fat and invaded the right lungs
General Thoracic and Cardiovascular Surgery (2022)
-
The many faces of solitary fibrous tumor; diversity of histological features, differential diagnosis and role of molecular studies and surrogate markers in avoiding misdiagnosis and predicting the behavior
Diagnostic Pathology (2021)
-
Solitary fibrous tumor of the greater omentum: case report and review of literature
Surgical Case Reports (2021)
-
Solitary fibrous tumor of the adrenal gland – its biological behavior and report of a new case
Surgical and Experimental Pathology (2021)