Abstract
Male breast cancer is poorly understood with a large proportion arising in the familial context particularly with the BRCA2 germline mutation. As phenotypic and genotypic differences between sporadic and familial male breast cancers have been noted, we investigated the importance of a hypoxic drive in these cancers as this pathway has been shown to be of importance in familial female breast cancer. Expression of two major hypoxia-induced proteins, the hypoxia-inducible factor-1α (HIF1A) and the carbonic anhydrase IX (CA9), examined within a large cohort including 61 familial (3 BRCA1, 28 BRCA2, 30 BRCAX) and 225 sporadic male breast cancers showed that 31% of all male breast cancers expressed either HIF1A (25%) and/or CA9 (8%) in the combined cohort. Expression of HIF1A correlated with an increased incidence of a second-major malignancy (P=0.04), histological tumor type (P=0.005) and basal phenotype (P=0.02). Expression of CA9 correlated with age (P=0.004) in sporadic cases and an increased tumor size (P=0.003). Expression of HIF1A was prognostic for disease-specific survival in sporadic male breast cancers (HR: 3.8, 95% CI: 1.5–9.8, P=0.006) but not within familial male breast cancer, whereas CA9 was only prognostic in familial male breast cancers (HR: 358.0, 95% CI: 9.3–13781.7, P=0.002) and not in sporadic male breast cancer. This study found that hypoxic drive is less prevalent in male breast cancer compared with female breast cancer, possibly due to a different breast microenvironment. The prognostic impact of HIF1A is greatest in sporadic male breast cancers with an alternate dominant mechanism for the oncogenic drivers suggested in high risk familial male breast cancers.
Similar content being viewed by others
Main
Male breast cancer is an uncommon and relatively uncharacterized disease entity. There is gathering evidence showing genetic and phenotypic differences and subsets distinct from female breast cancer.1, 2 Although male carriers of germline mutations in BRCA1 and BRCA2 and males from BRCAX families form a substantial proportion of male breast cancers, they are at a higher risk of developing breast cancer when compared with the general population,1 factors affecting tumorigenesis and phenotype are still relatively unclear but may be through distinct neoplastic pathways as is suggested by an emerging distinct BRCA2 phenotype of male breast cancer, characterized by a higher mitotic rate, higher grade, HER2 amplification, and an absence of PI3K pathway activation.1, 3, 4
In cancer, hypoxia is an important contributor to carcinogenesis5 and cancer phenotype.6, 7 Factors contributing to an imbalance of oxygen demand and supply lead to adaptive cellular mechanisms being initiated and involving vast transcriptional and post-transcriptional changes in gene expression, of which the hypoxia-inducible factor 1 (HIF1) is a master regulator.8 HIF1 is composed of two subunits; the HIF1α (HIF1A) and the aryl hydrocarbon receptor nuclear translator (ARNT), with HIF1A induced and continuously degraded via an association with the von Hippel–Lindau protein ubiquitin E3 ligase complex leading to degradation via the ubiquitin–proteosome pathway in normoxia.9 Under hypoxic conditions, however, HIF1A is translocated to the nucleus where it heterodimerizes with ARNT,10, 11, 12 which is constitutively expressed and independent of oxygen levels. The subsequent HIF1 complex regulates transcription of more than 100 target genes, and may account for >2% of all human genes either directly or indirectly.13 The result is an increase in erythropoietin, iron metabolism, angiogenesis, activation of the glycolytic pathway, activation of MAPK and PI3K signaling pathways promoting cell proliferation and survival and paradoxically apoptosis in some circumstances. Clinically, the HIF1A overexpression consequently correlates with poor prognosis, increased treatment resistance, and tumor associated mortality.14, 15
In female breast cancer, we have previously demonstrated that increased frequency of HIF1A expression is seen in BRCA1 type, basal-like cancers, and HER2 amplified female breast cancers,16 subsets that are infrequently seen in male breast cancer. Similarly, expression of the carbonic anhydrase IX (CA9) in female breast cancer has also been correlated with higher grade and poorer survival and is also overexpressed in triple negative (estrogen receptor negative, progesterone receptor negative, HER2 negative) tumors and associated with somatic loss of the BRCA1 protein.17 There is only one study of hypoxic markers in male breast cancer,18 comprising of 134 cases, which showed that 27% (34/125) and 7% (9/132) of cases demonstrated HIF1A and CA9 expression with HIF1A also associated with prognosis.18 The study was not stratified into sporadic and familial cancers, and currently there are no data in familial male breast cancer, or whether similar changes of the hypoxic pathway are present in particular intrinsic phenotypes as shown in female breast cancer.6 As both sporadic and familial male breast cancer patients also appear to have an increased predisposition to develop second malignancies, the effect or intrinsic response to hypoxia in these individuals may also be significant and has not been described in any study to date. Given the paucity of data in sporadic male breast cancer and absence of reports in the familial context, our aims were to (1) characterize HIF1A and CA9 levels in a large cohort of sporadic and familial male breast cancers, (2) correlate expression with conventional clinicopathological parameters and intrinsic phenotypes, (3) investigate expression in familial breast cancer stratified by carrier mutation status and (4) evaluate the prognostic significance of HIF1A and CA9 expression on disease-specific survival.
Materials and methods
Patient Accrual
An Australian based cohort of cases included familial male cases obtained from the KConFab resource (http://www.kconfab.org: criteria for admission to the kConFab study has been previously published)1 and sporadic male breast cancers obtained from the Peter MacCallum Cancer Centre and Melbourne Pathology. These cases were ascertained following a search of the relevant kConFab registry and pathology databases, and were diagnosed between 1980 and 2009 in Australia or New Zealand. Patients forming the Swedish cohort were identified through the Swedish National Cancer Registry.19 Males diagnosed between 1990 and 2007 within the Lund and Uppsala-Orebro regions that had available formalin-fixed paraffin-embedded tumor blocks, clinicopathological data and outcome data were included in the study. This work was carried out with approval from the Peter MacCallum Cancer Center Ethics Committee (Project No: 11/61) and the local ethics committee in Uppsala, Sweden (i2007/254), and the Lund University (2012/89).
Clinical parameters, including the American Joint Committee on Cancer 7th ed TNM staging, tumor recurrence, occurrence of non-breast primary tumors, and death were obtained from referring clinical centers, kConFab questionnaires and state death registries when available. Information on pedigree, mutational status and testing were available from the kConFab central registry. All available slides from cases were reviewed by a pathologist for relevant histopathological parameters. Histological classification was based on criteria set by the World Health Organization (2012).20
Germline BRCA1/2 Testing
Any Australian and New Zealand cases of male breast cancer with a strong family pedigree were referred to kConFab preceding this study. Mutation testing for BRCA1 and BRCA2 mutations was performed as reported previously1 on kConFab referred cases. Once the family mutation had been identified, all pathogenic (including splice site) variants of BRCA1 and BRCA2 were genotyped by kConFab in all available family member’s DNA. BRCAX cases were defined by cases with a strong family history meeting kConFab eligibility criteria (http://www.kconfab.org/Collection/Eligibility.shtml), but with absent BRCA1 or BRCA2 mutations within family members. In the Swedish cohort, only patients with a strong family history of breast and ovarian cancer had germline BRCA1/2 testing.
Tissue-Microarray Construction and Immunohistochemistry
Tumor tissue microarrays (1-mm cores), with a twofold redundancy, were prepared from archival formalin-fixed paraffin-embedded tissue blocks. Patient flow/use was as per the REMARK criterion21 is listed in Table 1. All 104 cases that were excluded were due to; blocks not being available, an absence of clinical and pathological information, or an absence of adequate material (ie, core biopsy diagnosis of breast cancer) for tissue-microarray construction.
Tissue-microarray sections were cut from each block at 4 μm thick intervals, dewaxed, placed through graded alcohol, and then into water. Antigen retrieval was performed using high pH EnVision FLEX Target Retrieval Solution (Dako) for 4 min at 124 °C for HIF1A and CA9. Staining for HIF1A (1:50 overnight incubation at 4 °C, Novus Biologicals) and CA9 (1:4000, 30 min at room temperature, Novus Biologicals) was performed using rabbit polyclonal antibodies. Antigen–antibody complex was detected using the Envision FLEX system (EnVision FLEX/HRP and EnVision FLEX DAB+Chromogen, DAKO) (Figure 1).
Scoring Criteria and Cut-offs
Scoring was performed according to a previously used semi-quantitative system.6, 16, 22, 23, 24 Briefly, HIF1A was scored only according to the presence (1+) or absence (0) of nuclear expression. Only tumors showing a strong membranous staining in ≥10% cells were considered positive for CA9.6
Statistical Analysis
Comparison of groups was made with using Mann–Whitney U for non-parametric continuous distributions and χ2- test for threshold data. Kaplan–Meier survival curves were plotted using breast cancer related death as the endpoint and compared using a log rank test. Analysis was performed with GraphPad Prism 5 software (GraphPad Prism version 5.04 for Windows, GraphPad Software, La Jolla CA, USA). A two-tailed P-value test was used in all analyses and a P-value of less than 0.05 was considered statistically significant.
Results
Expression of HIF1A and CA9 in Male Breast Cancer
In the overall combined sporadic and familial cohort, 25% (68/271) of cases were positive for HIF1A (Table 2). There was no statistically significant difference in HIF1A positivity between sporadic male breast cancers (28%, 59/213) compared with familial male breast cancers (16%, 9/58, P=0.06) with similar frequencies within the familial with BRCAX (14%, 4/28) and BRCA2 (15%, 4/27) males, and one of three (33%) BRCA1 male cancers. CA9 was less frequently observed in the overall combined cohort (8%, 22/276) (Table 2), with a higher proportion of positive cases in sporadic male breast cancers (9%, 19/218) when compared with familial male breast cancers (5%, 3/58, P=0.59); although numbers for BRCA1 are small, there was no statistically significant difference between BRCA1 (0%, 0/3), BRCA2 (7%, 2/28), and BRCAX (4%, 1/27) males.
In total, 31% (83/266) of all cases had expression of either marker with a higher frequency seen in sporadic male breast cancers (33%, 71/213) compared with familial male breast cancers (23%, 12/53, P=0.14) (Table 2), and the lowest frequency seen in BRCAX patients (19%, 5/26) (compared with BRCA1 (33%, 1/3) and BRCA2 (23%, 6/26) cohorts). Only seven cases had both CA9 and HIF1A positive expression, of which all were sporadic male breast cancers (7/213, 3% vs 0/53, 0% P=0.35).
Clinicopathological Correlation and Disease-Specific Survival.
HIF1A
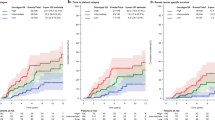
Overall in the combined sporadic and familial cohorts, HIF1A expression correlated with specific histological subtype (invasive carcinoma of no special type)(100 vs 90%, P=0.005) and basal cell intrinsic phenotype (100% 4/4, P=0.02) (Table 1a). Expression of HIF1A also correlated with a history of second-major malignancy (35 vs 22%, P=0.04) (Supplementary Table 2) but was not specific to a particular type of cancer. There was no correlation between HIF1A expression and patient age, tumor size, grade, stage or presence of lymph node, and distant metastasis (all P>0.05). There was no association between HIF1A expression and disease-specific survival (Figure 2a).
The association between HIF1A expression with invasive carcinoma of no special type (100 vs 92%, P=0.03) and basal cell intrinsic phenotype (100 vs 0%, P=0.02) was maintained within the sporadic male breast cancer cohort, as was the correlation with a history of second-major malignancy (37 vs 22%, P=0.04). In this group, HIF1A expression was associated with statistically significantly shorter disease-specific survival (HR: 3.8, 95% CI: 1.5–9.8, P=0.006) (Figure 3a).
Kaplan–Meier curves for disease-specific survival: (a) HIF1A expression in sporadic and familial male breast cancer, (b) CA9 expression in sporadic and familial male breast cancer, (c) HIF1A and/or CA9 expression—overall cohort, (d) HIF1A and/or CA9 expression in sporadic and familial male breast cancer.
Within familial male breast cancers, expression of HIF1A inversely correlated with a luminal intrinsic phenotype (67 vs 96%, P=0.02). Unlike in sporadic male breast cancer, a direct correlation between HIF1A nuclear expression and basal phenotype (1/9, 11% vs 0/49, 0%, P=0.17) was not seen, as only one case of basal phenotype was present within this cohort. There was no association between HIF1A expression and disease-specific survival (Figure 3a).
CA9
Overall, CA9 expression showed a correlation with older age (median 75.5 years vs 69.2 years, P=0.004) and an increased tumor size demonstrated by association with an advanced T-stage (stage 2–4) (81 vs 46%, P=0.003) (Supplementary Table 1b). Expression of CA9 was not prognostic for disease-specific survival (Figure 2b).
Within sporadic male breast cancers, expression of CA9 maintained a correlation with later age of onset (median 72.2 years vs 70.9 years, P=0.02) and an advanced T-stage (stage 2–4) (78 vs 48%, P=0.02). There were no other clinicopathological associations and no association with disease-specific survival (Figure 3b).
In familial male breast cancers, expression of CA9 correlated with a larger tumor size (median 26.7 mm (CA9 positive) vs 17.3 mm (CA9 negative), P=0.046) but no other clinicopathological factors. CA9 expression correlated with worse disease-specific survival (HR: 358.0, 95% CI: 9.3–13781.7, P=0.002) (Table 3, Figure 3b).
HIF1A and/or CA9
Overall, expression of any hypoxic marker (HIF1A and/or CA9) correlated with specific histological subtype (IC-NST)(99 vs 90%, P=0.010) and a basal cell intrinsic phenotype (100% 4/4, P=0.03) (Supplementary Table 1c). Expression of HIF1A also correlated with a history of a second-major malignancy (34 vs 21%, P=0.001). Prognostically, expression of any hypoxic marker was associated with a statistically significantly shorter disease-specific survival (HR: 2.2, 95% CI: 1.2–3.8, P=0.008) (Figure 3c, Table 3).
In sporadic male breast cancer, expression of any hypoxic marker correlated with a basal cell phenotype (100 vs 0%, P=0.04) and a history of second malignancy (37 vs 20%, P=0.01). Expression was also prognostically significant (Figure 2d) and associated with worse disease-specific survival (HR: 2.9, 95% CI: 1.5–5.8, P=0.002) (Figure 3d). In familial male breast cancer, there was no correlation with clinicopathological factors (Supplementary Table 1c) or disease-specific survival (Table 3, Figure 3d).
Discussion
This study is the first to examine the role of HIF1A and CA9 in a large cohort of sporadic and familial male breast cancers, including a small number of highly infrequent BRCA1 male breast cancers. Previous studies in female breast cancer show activation of the hypoxic pathway occurs early in tumorigenesis,25 promoting adaptive mechanism and conferring enhanced survival, invasive and metastatic capabilities. A strong geno-phenotypic association between pathway activation and BRCA1 associated tumors with a basal cell phenotype is seen in familial female breast cancers,16 and has yet to be comprehensively evaluated in male breast cancer. This study is the first to evaluate the effect of hypoxia is a cohort of male breast cancer segregated into familial and sporadic subgroups. All cases with a strong family history suggestive of an inherited predisposition were tested for BRCA1/2 germline mutation, the remainder of cases were categorized into ‘sporadic male breast cancers’. Although we cannot be entirely confident of the true sporadic nature, we believe the unparalleled large number of cases we present in the study, as the sporadic cohort should overcome the potential effect of any unknown germline mutation carriers, and thus contamination of the sporadic cases with hereditary cases should be minimal.
When compared with female breast cancer, the frequency of HIF1A expression is lower in all of our male breast cancer cohorts, being most marked is seen in familial male breast cancer, compared with familial female breast cancer16 (72, 38, and 41% of BRCA1, BRCA2, and BRCAX cases for female breast cancer compared with 33%, 14, and 15% in the respective male breast cancer cohorts). Similarly, sporadic female breast cancers6 showed a higher frequency of HIF1A positive tumors (50%) compared with sporadic male breast cancers in our study (28%), and comparable to the study by Kornegoor et al18 (27%). A similar but less extreme difference is seen when comparing CA9 expression in male breast cancer and female breast cancer,6, 26 again in keeping with the findings of Kornegoor et al.18 We also only found seven cases (3%) that expressed both HIF1A and CA9. Although this is below the range seen in female breast cancer (11–23%),6, 27 it is not unexpected given the decreased staining of either marker when compared with female breast cancer and the known variability in HIF1A and CA9 half-lives. HIF1A is rapidly degraded within minutes of oxygenation,28 whereas CA9 has a half-life of up to 38 h.29, 30
Importantly, using comparable staining and immunohistochemical scoring, our methodology has been closely addressed to minimize any potential differences to these comparable female breast cancer studies. Most of the staining observed was either ‘diffuse’ or ‘scattered’ for both markers, with no cases of perinecrotic staining seen mainly due to targeted sampling of non-necrotic areas for tissue-microarray assembly. Although the differentiation of staining patterns (perinecrotic vs diffuse) by Vleugel et al has been shown to be prognostically relevant and thought to correspond with greater activation of the hypoxic pathway, these parameters are not as consistently significant as the dichotomisation into expressing and non-expressing groups that we have used. Notably, although we also used a cut-off of ≥10% positive cells to define tumor CA9 positivity in an attempt to compare with female breast cancer studies, we found that all of our cases either showed completely absent staining or staining above this 10% threshold. We have also critically compared the use of tissue microarrays for the analysis of hypoxia markers in female breast cancer and found equivalent frequencies of expression with other studies that had used whole sections.31, 32 We feel this is a robust and well established methodology, and as our staining frequencies in male breast cancer are similar to Kornegoor et al,18 our detection rates were not substantially compromised.
Although there are low numbers of BRCA1 cases, reflecting the general paucity of these tumors within male breast cancer registries, our findings show that a decreased expression of hypoxia markers is characteristic of male breast cancers. This is in contrast to familial female breast cancer, which has an enhanced hypoxic drive.6 This may be due to a different breast microenvironment in males and may also be of critical importance in protecting male BRCA1 carriers from the development of breast cancers generally,6, 16, 25 and partly account for the low frequency of basal type in male breast cancer, even in BRCA1 associated tumors.1 Thus male breast cancer BRCA1 tumors that arise are likely to be through alternate mechanisms. The corollary of this is the observation that male breast cancers have a high frequency of hormone receptor expression. Thus, in female breast cancer, activation of the hypoxic pathway is associated with degradation of hormone receptor expression,33 but the low frequency of HIF1A in male breast cancers, enable retention of estrogen receptor and progesterone receptor.1
Nevertheless, overall male breast cancers that express HIF1A demonstrate a shorter disease-specific survival, as with female breast cancer, with a less clear association with the clinicopathological factors noted in female breast cancer. Interestingly in our study, when a male breast cancer had a strong identifiable driver, such as hypoxia in sporadic tumors or a predisposing BRCA2 mutation, there was a clear association with worse disease-specific survival. This perhaps suggests that high and low risk groups may exist in male breast cancers with prognostic relevance.
An interesting observation was the association between HIF1A expression and the occurrence of a second malignancy in both familial and sporadic male breast cancer. This is also novel and not previously described in female breast cancer. We and others have previously shown a high rate of second malignancies in patients with male breast cancer11, 34, 35 but the rates of second malignancies is significantly higher in HIF1A positive tumors (35%), prominent particularly in the prostate (Supplementary Table 2) and not dependent on BRCA2 status, and is higher than the range of reported rates of second malignancies (12–23%) in large male breast cancer series. It is unclear as to whether in these males, the effect of hypoxia is more significant due to inheritable modifiers or acquired lifestyle factors that may predispose to the onset of cancers.
Conclusion
Our findings suggest that oncogenic mechanisms resulting in overexpression of hypoxic markers are different between female and male breast cancer, and furthermore between sporadic and familial male breast cancer. Overall activation of the pathway is less frequently observed than in female breast cancer, and the association with BRCA1, basal-type tumors and HER2 amplification seen in female breast cancers is not clearly seen in male breast cancers within this study. The study also alludes to the prognostic importance of oncogenic drivers on disease-specific survival. As targeted therapies directed against HIF1A and downstream targets emerge,36, 37, 38 these observation may also have important therapeutic implications for identifying specific populations for screening and explaining treatment efficacy in familial male breast cancers.
References
Deb S, Jene N, Kconfab I et al. Genotypic and phenotypic analysis of male breast cancer shows under representation of the HER2 and basal subtypes in BRCA-associated carcinomas. BMC cancer 2012;12:510.
Johansson I, Nilsson C, Berglund P et al. Gene expression profiling of primary male breast cancers reveals two unique subgroups and identifies N-acetyltransferase-1 (NAT1) as a novel prognostic biomarker. Breast Cancer Res: BCR 2012;14:R31.
Ottini L, Silvestri V, Rizzolo P et al. Clinical and pathologic characteristics of BRCA-positive and BRCA-negative male breast cancer patients: results from a collaborative multicenter study in Italy. Breast Cancer Res Treat 2012;134:411–418.
Deb S, Do H, Byrne D et al. PIK3CA mutations are frequently observed in BRCAX but not BRCA2 - associated male breast cancer. Breast Cancer Res 2013;15:R69.
Harris AL . Hypoxia-a key regulatory factor in tumour growth. Nat Rev Cancer 2002;2:38–47.
Tan EY, Yan M, Campo L et al. The key hypoxia regulated gene CAIX is upregulated in basal-like breast tumours and is associated with resistance to chemotherapy. Br J Cancer 2009;100:405–411.
Vaapil M, Helczynska K, Villadsen R et al. Hypoxic conditions induce a cancer-like phenotype in human breast epithelial cells. PloS One 2012;7:e46543.
Wang GL, Jiang BH, Rue EA et al. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA 1995;92:5510–5514.
Srinivas V, Zhang LP, Zhu XH et al. Characterization of an oxygen/redox-dependent degradation domain of hypoxia-inducible factor alpha (HIF-alpha) proteins. BiochemBiophy Res Commun 1999;260:557–561.
Huang LE, Arany Z, Livingston DM et al. Activation of hypoxia-inducible transcription factor depends primarily upon redox-sensitive stabilization of its alpha subunit. J Biol Chem 1996;271:32253–32259.
Kallio PJ, Pongratz I, Gradin K et al. Activation of hypoxia-inducible factor 1alpha: post-transcriptional regulation and conformational change by recruitment of the Arnt transcription factor. Proc Natl Acad Sci USA 1997;94:5667–5672.
Vaupel P . The role of hypoxia-induced factors in tumor progression. Oncologist 2004;9 (Suppl 5):10–17.
Ke Q, Costa M . Hypoxia-inducible factor-1 (HIF-1). Mol Pharmacol 2006;70:1469–1480.
Dales JP, Garcia S, Meunier-Carpentier S et al. Overexpression of hypoxia-inducible factor HIF-1alpha predicts early relapse in breast cancer: retrospective study in a series of 745 patients. Int J Cancer 2005;116:734–739.
Generali D, Berruti A, Brizzi MP et al. Hypoxia-inducible factor-1alpha expression predicts a poor response to primary chemoendocrine therapy and disease-free survival in primary human breast cancer. Clin Cancer Res 2006;12:4562–4568.
Yan M, Rayoo M, Takano EA et al. BRCA1 tumours correlate with a HIF-1alpha phenotype and have a poor prognosis through modulation of hydroxylase enzyme profile expression. Br J Cancer 2009;101:1168–1174.
Neumeister VM, Sullivan CA, Lindner R et al. Hypoxia-induced protein CAIX is associated with somatic loss of BRCA1 protein and pathway activity in triple negative breast cancer. Breast Cancer Res Treat 2012;136:67–75.
Kornegoor R, Verschuur-Maes AH, Buerger H et al. Fibrotic focus and hypoxia in male breast cancer. Mod Pathol 2012;25:1397–1404.
Nilsson C, Koliadi A, Johansson I et al. High proliferation is associated with inferior outcome in male breast cancer patients. Mod Pathol 2013;26:87–94.
Lakhani SR, Ellis IO, Schnitt SJ et al. WHO classification of tumours of the breast. IARC: Lyon, 2012.
McShane LM, Altman DG, Sauerbrei W et al. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat 2006;100:229–235.
Boddy JL, Fox SB, Han C et al. The androgen receptor is significantly associated with vascular endothelial growth factor and hypoxia sensing via hypoxia-inducible factors HIF-1a, HIF-2a, and the prolyl hydroxylases in human prostate cancer. Clin Cancer Res 2005;11:7658–7663.
Couvelard A, Deschamps L, Rebours V et al. Overexpression of the oxygen sensors PHD-1, PHD-2, PHD-3, and FIH Is associated with tumor aggressiveness in pancreatic endocrine tumors. Clin Cancer Res 2008;14:6634–6639.
Soilleux EJ, Turley H, Tian YM et al. Use of novel monoclonal antibodies to determine the expression and distribution of the hypoxia regulatory factors PHD-1, PHD-2, PHD-3 and FIH in normal and neoplastic human tissues. Histopathology 2005;47:602–610.
Bos R, Zhong H, Hanrahan CF et al. Levels of hypoxia-inducible factor-1 alpha during breast carcinogenesis. J Natl Cancer Inst 2001;93:309–314.
van der Groep P, Bouter A, Menko FH et al. High frequency of HIF-1alpha overexpression in BRCA1 related breast cancer. Breast Cancer Res Treat 2008;111:475–480.
Vleugel MM, Greijer AE, Shvarts A et al. Differential prognostic impact of hypoxia induced and diffuse HIF-1alpha expression in invasive breast cancer. J Clin Pathol 2005;58:172–177.
Jiang BH, Semenza GL, Bauer C et al. Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am J Physiol 1996;271:C1172–C1180.
Rafajova M, Zatovicova M, Kettmann R et al. Induction by hypoxia combined with low glucose or low bicarbonate and high posttranslational stability upon reoxygenation contribute to carbonic anhydrase IX expression in cancer cells. Int J Oncol 2004;24:995–1004.
Turner KJ, Crew JP, Wykoff CC et al. The hypoxia-inducible genes VEGF and CA9 are differentially regulated in superficial vs invasive bladder cancer. Br J Cancer 2002;86:1276–1282.
Currie MJ, Hanrahan V, Gunningham SP et al. Expression of vascular endothelial growth factor D is associated with hypoxia inducible factor (HIF-1alpha) and the HIF-1alpha target gene DEC1, but not lymph node metastasis in primary human breast carcinomas. J Clin Pathol 2004;57:829–834.
van Diest PJ, Vleugel MM, van der Groep P et al. VEGF-D and HIF-1alpha in breast cancer. J Clin Pathol 2005;58:335 author reply 6.
Maity A, Sall W, Koch CJ et al. Low pO2 and beta-estradiol induce VEGF in MCF-7 and MCF-7-5C cells: relationship to in vivo hypoxia. Breast Cancer Res Treat 2001;67:51–60.
Marchal F, Salou M, Marchal C et al. Men with breast cancer have same disease-specific and event-free survival as women. Ann Surg Oncol 2009;16:972–978.
Ding YC, Steele L, Kuan CJ et al. Mutations in BRCA2 and PALB2 in male breast cancer cases from the United States. Breast Cancer Res Treat 2011;126:771–778.
Fox SB, Generali D, Berruti A et al. The prolyl hydroxylase enzymes are positively associated with hypoxia-inducible factor-1alpha and vascular endothelial growth factor in human breast cancer and alter in response to primary systemic treatment with epirubicin and tamoxifen. Breast Cancer Res: BCR 2011;13:R16.
Lundgren K, Holm C, Landberg G . Hypoxia and breast cancer: prognostic and therapeutic implications. Cell Mol Life Sci 2007;64:3233–3247.
Milani M, Harris AL . Targeting tumour hypoxia in breast cancer. Eur J Cancer (Oxford, England: 1990) 2008;44:2766–2773.
Acknowledgements
We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow up Study (funded 2001–2009 by NHMRC and currently by the National Breast Cancer Foundation and Cancer Australia #628333) for their contributions to this resource, and the many families who contribute to kConFab. kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC, and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania, and South Australia, and the Cancer Foundation of Western Australia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Deb, S., Johansson, I., Byrne, D. et al. Nuclear HIF1A expression is strongly prognostic in sporadic but not familial male breast cancer. Mod Pathol 27, 1223–1230 (2014). https://doi.org/10.1038/modpathol.2013.231
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2013.231
Keywords
This article is cited by
-
Public transcriptome database-based selection and validation of reliable reference genes for breast cancer research
BioMedical Engineering OnLine (2021)
-
Understanding metabolic alterations and heterogeneity in cancer progression through validated immunodetection of key molecular components: a case of carbonic anhydrase IX
Cancer and Metastasis Reviews (2021)
-
Activation of the hypoxia pathway in breast cancer tissue and patient survival are inversely associated with tumor ascorbate levels
BMC Cancer (2019)
-
miRNA profile of neuroprotection mechanism of echinomycin in Parkinson’s disease
Molecular & Cellular Toxicology (2017)
-
Overexpression of HIF1α and CAXI predicts poor outcome in early-stage triple negative breast cancer
Virchows Archiv (2016)